Advancements of paper-based sensors for antibiotic-resistant bacterial species identification

Introduction
Bacterial infections pose a significant threat to global public health, standing as the second leading cause of mortality worldwide. These infections, ranging from common ailments to life-threatening conditions, contribute to a substantial burden on healthcare systems and economies across the world. Per the Institute of Health Metrics and Evaluation – Global Burden of Diseases 2019 report, about 13.7 million deaths were associated with infections, of which nearly 7.7 million (estimating about 1 in 8) deaths were linked to bacterial infections1,2. Bacterial infections are predominantly associated with 33 bacterial species, including the highly virulent ESKAPE pathogens, namely Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species3,4,5. These pathogenic bacteria are responsible for 75% of deaths caused by infections in the lower respiratory tract, blood, peritoneal, or intra-abdominal areas1. With the emergence of antimicrobial resistance, the challenge of combating bacterial infections has become even more daunting. Despite advances in medical science, bacterial pathogens continue to evolve and adapt, rendering traditional treatments ineffective in numerous cases. Antimicrobial-resistant (AMR) bacterial species pose a formidable global health threat, marked by their swift dissemination and the substantial economic pressure they exert on healthcare systems, leading to prolonged hospitalizations, increased healthcare costs, and elevated mortality rates6. In the United States alone, they cause more than 2 million infections and lead to 23,000 deaths annually7. Worldwide, AMR jeopardizes advancements in healthcare, food production, and ultimately, life expectancy. The widespread overuse and misuse of antibiotics in both clinical and agricultural settings over the past few decades has accelerated the emergence and spread of resistant strains, giving rise to an epidemic of AMR4,5. AMR bacteria are ubiquitous in the community and can be acquired through various routes, including nosocomial infections, post-surgery complications, and contaminated food sources8,9,10. Furthermore, infections caused by resistant bacteria, such as those leading to sepsis, exhibit a mortality rate of 35%, emphasizing the gravity of the antibiotic resistance crisis11. Managing septic patients is a critical clinical challenge, necessitating prompt and effective antibiotic prescriptions, which demand rapid diagnosis of resistant infections and subsequent antibiotic susceptibility testing (AST)12,13. As such, the prevention, diagnosis, and management of bacterial infections remain critical priorities in healthcare and biomedical research. Efforts to address this challenge include the development of novel antimicrobial agents, improved diagnostic tools, and innovative treatment strategies aimed at curbing the spread of bacterial pathogens and reducing the associated morbidity and mortality. A multifaceted approach encompassing prudent antibiotic usage, infection prevention strategies, and novel therapeutic interventions is essential in mitigating this formidable threat.
Antimicrobial resistance and tools for detection
AMR is the ability of a microorganism to survive after standard treatments with antimicrobial agents. Bacterial species may exhibit resistance to antibiotics through intrinsic or acquired mechanisms14. Intrinsic resistance refers to the natural, inherent ability of a bacterial species to resist the effects of an antibiotic due to its structural and/or functional characteristics. This type of resistance is genetically encoded in the bacterial chromosome and does not arise from recent genetic alterations or horizontal gene transfer. Intrinsic resistance includes permeability barriers, efflux pumps, and lack of targets15. Acquired resistance refers to the ability of bacteria to develop resistance to an antibiotic to which they were previously susceptible16. This can occur through several mechanisms, including horizontal gene transfer processes such as conjugation, transformation, or transduction and mutations in the bacterial genome. These processes alter the target site of the antibiotic, reduce drug uptake, or increase drug efflux, leading to resistance17,18. S. aureus, for example, developed resistance to penicillin and other β-lactams, which inhibit bacterial cell wall biosynthesis, ultimately leading to cell lysis and death. Specifically, β-lactam antibiotics target penicillin-binding proteins (PBPs) and acylate a catalytically essential serine residue with the reactive β-lactam core, yielding stable acyl-enzyme complex (AEC)19. This prevents the PBP from catalyzing the transglycosylation and transpeptidation reactions critical for the biosynthesis of peptidoglycan20,21. Enzyme-mediated resistance to β-lactam antibiotics is driven by β-lactamases, enzymes synthesized by both Gram-positive and Gram-negative bacteria, which catalyze the hydrolysis of the β-lactam amide22. Notable examples of hydrolyzing enzymes produced by various bacteria include oxacillinases, cephalosporinases, carbapenemases, acetyltransferases, adenyltransferases, and phosphotransferases23. Other mechanisms of resistance involve genetic mutations affecting the antibiotic target site. For example, β-lactams target the PBPs within the cell wall of S. aureus. Consequently, various strains of S. aureus have acquired resistance to this class of antibiotics due to genetic mutations in the PBPs, leading to a reduced effectiveness of β-lactams24. Bacterial species frequently acquire drug resistance genes and clusters, such as Staphylococcal Cassette Chromosome mec (SCCmec) or vancomycin resistance gene (van), through horizontal gene transfer, thereby diminishing the effectiveness of antibiotics by reducing their binding capability25,26,27. Bacterial species have developed resistance to different antibiotic classes, such as aminoglycosides, fluoroquinolones, or oxazolidinone by various mechanisms, including enzymatic modifications or genetic alterations of the binding sites25. Overexpression of efflux pumps limits the susceptibility of bacteria to antibiotics28. This is commonly observed in species like Acinetobacter baumannii and Helicobacter pylori, particularly with antibiotics from the tigecycline, carbapenem, or clarithromycin classes29. Bacterial species also exhibit porins, which are β-barrel proteins that traverse cellular membranes and function as channels, facilitating the passive transport of hydrophilic drugs into the cells30. Two primary mechanisms of antibiotic resistance based on porins have been documented in clinical isolates: changes in the membrane profiles, characterized by either the loss or significant reduction of porins, or the substitution of one or two major porins with others, and altered porin function resulting from specific mutations that decrease permeability30. For example, A. baumannii reduces the expression of porin proteins, thus developing resistance to antibiotics like carbapenems14,29.
The dynamics of the emergence of AMR species overtime is illustrated in the Fig. 1. The first sign of resistance was observed in S. aureus to penicillin in the early 1940s. Subsequently, over the next 60–70 years, multiple bacterial species have acquired resistance to various classes and generations of antibiotics.

MALDI TOF matrix-assisted laser desorption/ionization-time of flight, FISH fluorescence in situ hybridization, MBT-ASTRA matrix-assisted laser desorption ionization Biotyper antibiotic susceptibility test rapid assay, PCR polymerase chain reaction.
The serendipitous discovery of penicillin by Alexander Fleming in 1928 marked a turning point in medical history, introducing the antibiotic era and revolutionizing the treatment of bacterial infections7,31. Nevertheless, even in the initial phases of antibiotic application, Fleming discerned the emergence of bacterial populations displaying resistance to penicillin, hinting at the looming challenge of antibiotic resistance32. Prior to the therapeutic introduction of penicillin a few years, a bacterial penicillinase was identified, signaling an early recognition of mechanisms that could potentially compromise its efficacy33. This pivotal discovery prompted the medical community to seek effective ways to quantify antibiotic efficacy, leading to the establishment of standardized antimicrobial susceptibility testing methods during the mid-20th century34. From the 1940s to the 1960s, pioneering techniques such as broth dilution, agar dilution, and disk diffusion assays became essential for determining the minimum inhibitory concentrations (MICs) or zones of inhibition for bacterial growth35,36,37,38,39. While accurate, these methods were labor-intensive and time-consuming, limiting their efficiency. The 1970s and 1980s saw a shift towards automation, with the introduction of innovative instruments like Autobac disc elution system40,41, Etest42, AutoMicrobic, Vitek43,44, and MicroScan45, which offered high-throughput testing (Fig. 1). Despite advancements in automation, these techniques still relied on isolated cultures and required overnight incubation, delaying real-time analysis and decision-making. The advent of molecular techniques in the 1980s through 2000s, including polymerase chain reaction (PCR)46, DNA microarrays, and genomic sequencing47,48, marked the onset of the molecular era in AST49. These techniques allowed rapid profiling of antibiotic resistance genes, providing insights into the genetic basis of resistance mechanisms. However, a critical limitation was the lack of direct phenotypic correlation at the functional level, necessitating confirmatory culture-based testing. In the contemporary era, spanning from the 2000s to the present day, remarkable progress in advanced imaging50, microfluidics, and nanoscale sensors51,52 was observed, enabling faster quantification of phenotypic AST46,53,54,55,56. Techniques like simplified blood culture system, plasmonic imaging and tracking, single bacterial cell analysis, micromotion tracking, and morphotyping hold great promise for delivering rapid and accurate results10,57,58,59,60. Nevertheless, challenges related to standardization, regulatory approval, and infrastructure requirements have hindered their widespread clinical adoption.
The Clinical Laboratory Standards Institute (CLSI) M100 has developed standards for the identification of AMR61. Per these standards, resistance to methicillin is declared when oxacillin MIC > 4 µg/mL, and for vancomycin-intermediate strains 4–8 µg/mL. Using the disk diffusion test, MRSA strains show zone diameter ≥17 mm, while VISA strains show ≥ 15 mm61. Though accurate, these techniques lack rapid diagnoses and require ~2–3 days for determining the presence of AMR bacterial species. These techniques have limited potential as a sensitive, specific, and point-of-care testing (POCT) platform62.
Throughout history, AST has evolved in response to the mounting threat of antibiotic resistance. By learning from the past and embracing innovative approaches and technologies, we can strive for more effective antibiotic stewardship and forge a path toward a future where antibiotics remain potent tools in the ongoing battle against resistant infections. The outlook for AST is promising, with the potential integration of genotypic and phenotypic techniques, leveraging sequencing and rapid culture-independent phenotyping. Microfluidic technologies aimed at pathogen isolation from complex samples, as well as multiplexed assays, have the potential to streamline the testing process and enable direct analysis from patient specimens. Furthermore, the development of POCT platforms might empower healthcare providers with timely and tailored treatment decisions, optimizing antibiotic stewardship.
Point-of-care testing
POCT are developed to diagnose infections rapidly and accurately, and to monitor diseases where the treatment facilities are not easily available. POCT are any clinical diagnostic tools that can be used at the time and location of the patient instead of a central laboratory. In consideration of resource-limited settings, the World Health Organization Special Program for Research and Training in Tropical Disease (WHO/TDR) in 2003 developed an ASSURED criteria for POCT. For this, the tests must be affordable, sensitive, specific, user-friendly, robust and rapid, equipment-free, and deliverable to consumers63,64. These criteria are designed as a benchmark for diagnosis of major pathogens causing infections including AIDS, syphilis, and tuberculosis63. However, tests developed using ASSURED criteria lack the ability to scale up in resource-limited settings. Therefore, real-time connectivity and ease in sample collection were added to the existing ASSURED criteria to provide a new criterion termed REASSURED. Several diagnostic tools using the ASSURED criteria have been developed for the identification of pathogens like human immunodeficiency virus, plasmodium parasites, and bacteria like Mycobacterium tuberculosis and Treponema pallidum65. Additionally, POCT has also been developed for the diagnosis and monitoring of diseases such as diabetes mellitus and hypertension. The ability of patients to self-monitor or rapidly identify disease conditions can ultimately improve treatment strategies and overall public health. Therefore, the advancement of POCT in the medical and pharmaceutical fields has great importance.
POCT utilizing biosensing tools is emerging to bridge the gap between the biological and electronic worlds. Sensors are analytical devices that measure responses from biological components and convert them into digital signal outputs. A sensor typically consists of four components: a sample or analyte of interest, recognition elements or receptors, transducers or detection methods, and signal process (Fig. 2). The recognition element governs the specificity of the tool, while the detector determines the sensitivity. The interaction between these two components involves the generation of signals that are then processed into a quantifiable output. Sensors can be classified depending on the target (antibodies, nucleic acids, enzymes, or whole microbial cells) or the type of transducer (electrochemical, optical, ion-sensitive, thermal, and resonant)66. Electrochemical sensors use electrodes for the detection of the analyte67. Electrochemical sensors can be further subcategorized into amperometric sensors that measure change in current, potentiometric sensors that measure changes in charge, and impedimetric sensors that measure changes in conductance67. In contrast, optical sensors detect optical changes upon interaction with analytes. Optical sensors include the detection of analytes based on absorbance, fluorescence, chemiluminescence, surface plasmon resonance, and surface-enhanced Raman spectroscopy68. Thermal or calorimetric sensors detect temperature changes to identify analytes while resonant sensors utilize mass changes of analytes upon interaction with the recognition elements69.

Sensing tools are comprised of four components: (1) Samples such as bacteria presented in biological sources (eukaryotic cells, human specimens, food or environment); (2) Sensing receptors, i.e., antibodies, nucleic acids, enzymes that interact with the bacterial targets; (3) Detectors, which are measuring the interaction between the sample and receptors as a function of electrochemical, optical, ion, thermal, or resonance changes; and (4) Signal processing, a component converting the signals from the detector into measurable output.
Sensing tools have been developed for a wide variety of applications. As diagnostic tools, biosensors have been created to diagnose cancer70, neurogenerative diseases71, and cardiac diseases72 based on the presence of specific biomarkers. Additionally, sensors for the diagnosis and monitoring of autoimmune diseases73, diabetes74, and arthritis75 have been developed66. Moreover, the identification of infection-causing pathogens such as bacteria, viruses, and unicellular parasites has significantly improved with the usage of sensing tools76. Beyond disease diagnosis, biosensors for monitoring the quality of food, air, water, and environmental samples have also been developed77. Sensing tools are typically designed with the goal of requiring low sample volumes, being instrument-free and portable. They are inexpensive, can be easily miniaturized, sometimes designed to be wearable, and are user-friendly. Therefore, sensors offer advantages over conventional diagnostic tools and can be developed as POCT with an emphasis on settings with limited resources.
Paper-based sensors
Paper as a platform for facilitating chemical analysis has gained popularity because of its affordability, portability, cost-efficiency, ease of use, as well as hydrophilicity. The use of a paper-based sensor was first documented in the 1600 s, with litmus paper employed for measuring the pH of solutions. The applicability of paper in analytical chemistry expanded upon the invention of paper chromatography78. Paper test strips or dipsticks were developed in the 1950s for detecting blood glucose levels and for pregnancy testing79. Over the years, the use of paper-based sensors as POCT in disease diagnosis has seen significant advancements. Beyond disease diagnosis, paper-based sensors have been used for monitoring the environment, food safety testing, as well as bacterial and viral pathogen detection80,81,82. Paper-based sensing platforms fulfill the REASSURED criteria due to their unique morphological properties. The cellulose matrix and hydrophilic nature of paper allow for the easy wicking of samples and for the use of aqueous solutions for analysis. The morphology and porous structure allow the immobilization of biological, organic, as well as inorganic reporters. Additionally, paper can be functionalized to meet the desired requirements. For example, the hydrophilicity, reactivity, and permeability can be altered. Lastly, paper is easily and inexpensively available worldwide; it is biodegradable and biocompatible. Therefore, paper, as a solid platform for analytical devices has advantages over conventional techniques.
Paper-based analytical devices using microchannels were developed in the early 2000s technique for glucose detection using urine sample83. Microfluidic paper-based analytical devices are now being investigated as tools for disease diagnostics, as well as for the detection of pathogenic bacteria and viruses82. These paper-based platforms can be designed to be compatible with absorbance, fluorescence, and electrochemical detection of analytes. A wide variety of papers like Whatman filter paper (grades 1–4), chromatography paper, filter paper, etc. have been chosen for developing paper-based sensors. The choice of paper depends on its porosity, liquid retention, and flow rate. For example, cellulose-based Whatman papers show an increase in pore size and retention from Grades 1 to 4. Conversely, papers based on nitrocellulose are relatively hydrophobic and possess uniform pore sizes.
Many paper-based platforms have been developed for the sensing of pathogenic bacteria. A comprehensive review by Mazur et al. highlights the current advances of paper-based approaches for the identification of bacterial species84. Paper-based platforms for sensing pathogenic bacteria include paper-based ELISA, lateral flow strips, paper strips, paper-based analytical devices (PADs), microPADs, paper microzone plates, paper-based LAMP devices, and paper discs84. Table 1 summarizes the main paper-based devices reported to date for the identification of AMR species categorized by the detection techniques: colorimetric, fluorometric, and photoelectrochemical analysis. Along with targets and receptors used in the detection of bacterial species, the table highlights their overall limit of detection (LOD) and analysis time. All these sensors are fabricated using different techniques like wax patterning, wax printing, ink-jet printing, photolithography, chemical deposition, or laser cutting. Liana et al. and Bandopadhyay et al. have extensively described all fabrication methods for paper-based sensors85,86. In brief, fabrication techniques like wax patterning or wax printing use hydrophobic wax to create the patterns of microfluidic channels85. Alternatively, photolithography utilizes photochemical polymerization to create microfluidic channels using photomasks85. Overall, these fabrication techniques enable compounds to penetrate through the depth of the paper and create hydrophobic barriers, while maintaining the hydrophilicity of the rest of the paper.
The development of AMR species over time and the lack of rapid and accurate identification of the species has an impact on public health. Therefore, POCT has been widely investigated as an aim to detect AMR species. Paper-based sensors represent a promising group of POC diagnostic tools as they offer rapid, cost-effective, user-friendly, and, in some cases, equipment-free ways of identifying bacterial species. Due to the unique morphological and structural properties of paper, it can efficiently support various analytical methods that measure signals using colorimetric, fluorometric, and electrochemical detection. Paper-based platforms for antimicrobial sensing offer advantages as point-of-care tools by providing lower detection limits, requiring smaller sample volumes, and enabling shorter analysis times. As illustrated in Fig. 3, this review delves into a variety of paper-based platforms used to identify AMR bacterial species, emphasizing targets such as whole bacterial cells, nucleic acids, and enzymes. It also explores the use of nanosized particles for identifying drug-resistant bacteria.

Paper-based sensors using detection methods (colorimetry, fluorometry, surface-enhanced Raman spectroscopy, and nanoparticles) and various targets for identification of AMR bacterial species.
Paper-based techniques for AMR species identification
Colorimetric detection
Colorimetric sensors exhibit visible color changes in response to a positive interaction with the analyte. In this case, the transducer is typically a molecule or a molecular fragment capable of changing its spectral properties, such as absorbance maximum and/or intensity, when the recognition element interacts with the analyte. Bacterial sensing using colorimetric detection, chromogenic substrates as pH indicators, enzyme-catalyzed reactions, or nanoparticle aggregation/accumulation is widely used87. These chromogenic substrates change colors depending on changes in pH, metabolic and/or enzymatic activities, or due to the interaction of nanoparticles with the target site87. The development of AMR in bacterial species is manifested by phenotypical changes that alter the targeting site for antibiotic susceptibility, metabolic activity, and the presence of inactivating enzymes in the bacterial species88. Taking advantage of these changes, colorimetric detection using whole bacterial cells, enzymes, and nucleic acids has been developed for sensing AMR bacteria.
Whole cells
Bacterial species show diversity in terms of their size, shape, and morphology89. They typically range from 0.5 µm to 5 µm in size, and can exist in various forms such as spherical, rod-like, spiral, and comma shapes90. They are commonly classified as Gram-positive and Gram-negative based on their Gram-staining properties. Gram-positive bacteria possess a thick peptidoglycan layer and lack an outer membrane, whereas Gram-negative bacteria have a thin peptidoglycan layer surrounded by an outer membrane containing lipopolysaccharides91. These structural differences influence the composition of the surface proteins in the bacterial species, thereby providing distinctive targets for sensing. In addition, AMR bacterial species might exhibit altered morphological features compared to susceptible ones18,25. For instance, the development of resistance to methicillin and/or vancomycin in S. aureus causes thickening of the cell wall and septum92. Therefore, whole cell-based sensing focuses on the identification of bacteria by interacting with specific targets within the cell envelope, such as cell surface proteins and receptors, peptidoglycan, lipo- and exopolysaccharides, extracellular DNA (eDNA) or enzymes89.
Observation of bacterial growth dynamics in the presence of antibiotics remains the most direct method of AST. Accordingly, paper-based platforms such as paper-based AST chips have been developed to test antibiotic susceptibility93,94. These devices streamline the traditional disc diffusion test, allowing real-time observation of changes in bacterial concentrations. Paper-based microfluidic devices are fabricated using a wax printing method on chromatography paper. In this method, wax is applied to the paper’s surface and then melted on a hot plate, causing the wax to penetrate through the paper’s porous structure and create hydrophobic barriers95. The wax also forms hydrophobic barriers around the hydrophilic microfluidic channels that form test zones designed as flower petals with center as sample loading zone. Each test zone immobilizes and dries the antibiotic of interest at increasing concentrations, along with a chromogenic substrate, facilitating easy and effective analysis. A chromogenic substrate is a colorless chemical that changes color in the presence of an enzyme. For example, resazurin, a redox indicator, can be used as a chromogenic substrate to identify metabolically active bacteria96. Active bacteria internalize resazurin, a blue dye, and reduce it to resorufin, which is pink, in the presence of intracellular diaphorase enzymes97,98. This color change provides a visual signal identifying bacteria that are not affected by antibiotics. Samples containing bacterial species are loaded in the center of the device and then radially diffuse into the test zones. Bacteria that are metabolically active or unaffected by antibiotics change color due to the redox reaction mediated by the bacterial enzymes. This paper-based AST device is a robust, user-friendly technique that requires low sample volumes to identify antibiotic-resistant strains of bacteria such as E. coli, K. pneumoniae, and A. baumannii against several antibiotics, including gentamicin, ciprofloxacin, ampicillin, and meropenem93.
To improve the previous design where only a single antibiotic was immobilized per chip, a multiplex paper-based microfluidic device called bacterial paper AST chip (Bac-PAC) was fabricated. In this layout, multiple antibiotics were added to the same chip with chromogenic substrates like resazurin or 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-carboxanilide-2H-tetrazolium (XTT) dye (Fig. 4A). Similarly to resazurin, XTT is used as an indicator for viable cells, wherein a metabolically active bacteria it is converted to formazan (orange color)99. This device allows for simultaneous testing of multiple antibiotics, increasing efficiency and providing more comprehensive resistance profiles. In contrast to the paper-based AST chip taking 14–16 h, Bac-PAC identified bacterial species within 10 h94. This improved sensor showed 90% accuracy in identifying 12 different AMR bacterial species, reduced cost and analysis time, and improved shelf life94.

A Whole cells that interact with chromogenic substrates in metabolically active bacteria, indicated the absence of AMR. Reprinted (adapted) with permission from ref. 94. Copyright {2022} Springer Nature; and B Enzymes like β-lactamases that are present in AMR bacterial species show changes in color of chromogenic. Reproduced (adapted) with permission from ref. 106. Copyright {2017} John Wiley and Sons.
Surface proteins can also be used as targets for whole-cell-based sensing. In this approach, recognition elements such as antibodies or aptamers are needed, often integrated within microfluidic systems, to specifically bind and identify bacteria based on their surface markers100. Various paper-based sensors were fabricated using a laser-based direct-write procedure. This technique allows to produce three-dimensional (3D) paper structures101. In this technique, Whatman filter papers are stacked to form the device which consists of three layers: a bottom layer containing chromogenic agar for bacteria identification, a middle layer containing the sample injection port, and a top layer containing an array of antibiotics for susceptibility testing. While currently limited to the identification of E. coli, this technique enables simultaneous detection of bacteria and conducts AST. Identification of bacterial species is performed by growing the species on paper-based chromogenic agar plates. At the same time, AST takes place as the microfluidic channels transport the samples to the laser-printed zones containing the array of antibiotics. Therefore, compared to traditional techniques for AMR identification, this method provides a rapid, user-friendly, and convenient tool for AMR detection. These devices require relatively few experimental steps, allow for direct visualization of AMR species presence, and are easy-to-use as a POCT.
Enzymes
Specific enzymes and enzymatic reactions can serve as unique targets in the process of identifying bacterial species. During growth bacteria produce and secret a variety of enzymes essential for their fitness and functionality, including enzymes involved in cell wall biosynthesis, protein and nucleic acid synthesis and processing, as well as other metabolic processes23. Therefore, inhibiting the activity of specific enzymes becomes a useful strategy to achieve a bactericidal effect. For instance, β-lactams target PBPs to inhibit cell wall synthesis, while drugs in the quinolone class target enzymes such as DNA gyrase to inhibit DNA replication and, consequently, cell proliferation102. However, to overcome these bactericidal effects, bacteria produce inactivating enzymes to prevent the effect of antimicrobial agents. Inactivating enzymes include hydrolases, transferases, and oxidoreductases. Hydrolases, such as β-lactamases, hydrolyze β-lactams rings present on the antimicrobial agent, thus inhibiting their action on PBPs22,103. Transferases inactivate the action of antimicrobial agents by transferring different chemical moieties onto antimicrobial agents, thus altering their specificity and targetability to the bacteria103. Lastly, oxidoreductases inactivate antibiotics by transferring electrons from an oxidizing group to a reducing group104. Therefore, these modifying enzymes are widely investigated as targets for predicting antimicrobial resistance. Taking advantage of the presence of these inactivating enzymes in bacterial species, inactivating enzymes are now being used as targets for the identification or sensing of AMR species.
Paper-based microanalytical devices (µPADS) such as paper discs have been developed for AMR species identification using enzymes as targets105,106,107. These paper discs, made of Whatman filter paper, contain chromogenic substrates for detecting metabolically active bacterial species105. Bacteria alter their enzyme levels in their dormant and active forms. For example, E. coli increases its oxidoreductase levels in a dormant state, and bacterial dormancy can be directly correlated with antibiotic resistance108. Therefore, metabolically active and dormant bacteria can be differentiated by analyzing enzymes like oxidoreductase. Chromogenic substrates like tetrazolium salts have been used to differentiate between these two bacterium types. The presence of oxidoreductases is marked by the development of a purple color owing to the reduction of tetrazolium salts to formazan derivatives. For the identification of other AMR bacterial species like S. aureus, µPADS fabricated with the wax printing method have been reported107, where enzymes such as β-galactosidases, alkaline phosphatases, and β-lactamases are targeted. Chromogenic substrate CPRG (chlorophenol red-β-d-galactopyranoside), initially yellow, readily penetrates the bacterial cell membranes and is degraded by β-galactosidases to produce chlorophenol, resulting in a red coloration109,110. Alkaline phosphatases are excreted by the bacteria and can be detected using chromogenic substrates like BCIP (5-bomo-6-chloro-3-indolyl phosphate p-toluidine salt) and nitrophenyl phosphate105,111. In the presence of alkaline phosphatases, colorless BCIP is hydrolyzed to form a magenta-colored product, while nitrophenyl phosphate is cleaved to produce a yellow color. β-Lactamases, in their turn, are identified with nitrocefin, which changes from yellow to red in color in the presence of the enzyme (Fig. 4B)112. A novel chromogenic substrate, HMRZ-68, has recently been developed for the detection of extended-spectrum β-lactamases113. It has also been investigated for the identification of AMR bacterial species114.
Along with paper discs, a nanoelectrokinetic paper-based analytical device has been developed for AMR bacterial species identification115. In this device, bacterial cells are lysed using an external voltage, which causes the release of enzymes like β-lactamase. The reaction of these enzymes with nitrocefin in a nanoporous membrane causes the nitrocefin patch to change its color from orange to red.
Thus, enzymes causing modification of antibiotic agents have been widely investigated for the identification of AMR species. The immediate reaction of the colored substrates with enzymes provides a rapid identification of the species. Paper discs with pre-adsorbed chromogenic substrates can provide a state-of-the-art technology with consideration to resource-limited settings.
Nucleic acids
Genetic mutations and horizontal gene transfer are pivotal in driving the emergence and spread of drug resistance among bacterial species by altering target sites and thereby reducing the efficacy of antibiotics. For example, methicillin, a β-lactam antibiotic, inhibits bacterial cell wall synthesis by targeting the PBPs. However, methicillin-resistant S. aureus (MRSA) strains exhibit a high level of resistance to β-lactams due to the acquisition and expression of PBP2a, an alternate PBP encoded in mecA gene located on a mobile genetic element known as Staphylococcal Cassette Chromosome mec (SCCmec)116,117. Similarly, S. aureus develops resistance to vancomycin, a glycopeptide antibiotic, due to the acquisition of vanA gene from vancomycin-resistant enterococci118. This gene modifies the peptidoglycan layer, thus reducing the affinity of vancomycin119. Therefore, molecular techniques for the rapid identification of resistance genes are essential for the detection of AMR species120. Genotypic analysis is one of the most reliable approaches to AST when assessing resistance linked to known genetic markers. Nucleic acid-based methods target DNA or RNA to detect and identify pathogens. This detection method has been gaining an increased interest as it can be designed to target specific genes or sequences.
One of the popular techniques that use nucleic acids for the identification of AMR bacteria is loop-mediated isothermal amplification (LAMP). LAMP is a detection method used in POCT for the identification of bacteria121. Similar to PCR, LAMP amplifies target nucleic acids without using multiple thermal cycles and maintains a constant temperature of about 55–60 °C122. Compared to traditional techniques like PCR, this technique is robust as it can rapidly detect fewer copies of the gene of interest while at a constant temperature. The LAMP assay utilizes four to six specific primers designed to bind to distinct regions of the DNA, facilitating the amplification of the target sequences. LAMP reactants contain DNA-staining dyes that help in the colorimetric analysis123. Paper-based LAMP sensors with colorimetric detection have been developed as rapid and cost-effective devices to identify specific DNA124. These multi-layered sensors are fabricated using a wax-patterning method. The gene of interest is isolated from the bacterium and loaded onto the center paper strip containing the LAMP reagents and chromogenic substrates. These substrates aid in the visual identification of the target gene. Chromogenic substrates like silver nanoplates have been used for the visual identification of MRSA species124. Metals like silver and gold show higher binding affinities to nucleic acids. They exhibit surface plasmon resonance (SPR)—a phenomenon causing resonance of free electrons generated because of light hitting a metal surface125. Due to this phenomenon, they are able to change their color in the solution to red. Therefore, this method offers a rapid (30 min) and sensitive (1 copy of the gene) tool for the identification of MRSA species124.
In conventional LAMP reactions, new DNA synthesis releases protons in the solution, but due to high buffer concentrations (greater buffer capacity), the pH of the solution is not altered. At the same time, a lower buffer concentration combined with phenolphthalein-treated discs can be used for efficient visualization112. Phenolphthalein is a pH indicator that is pink in color when in a solution with alkaline pH, and turns colorless in acidic or neutral solutions. This technique has been used in the detection of of vanA and vanB genes present in vancomycin-resistant Enterococcus species. The production of AMR genes in LAMP reactions changes the pH of paper, which is indicated by a change in the color of the discs from pink to colorless. Overall, this technique identifies vancomycin-resistant Enterococcus in 45–70 min and requires a low sample concentration (102 CFU/mL)112.
Fluorometric detection
Colorimetric analysis is the simplest technique for detecting analytes, as the signal can be detected visually, often without the need for any instrumentation at all. At the same time, fluorimetry presents an important advantage: while keeping the simplicity of measurement, it is comparatively more sensitive than colorimetry due to the higher contrast of even the weakest signal.
LAMP that is successfully used in colorimetric analysis can also be combined with fluorescent reporters to visualize the signal. For example, in a paper-based LAMP device, a cellulose membrane sandwich between two pieces of adhesive tape is used as a reaction pad for the identification of bacteria126. The cellulose membrane is coated with streptavidin and immobilized with a biotinylated primer specific to MRSA. Subsequently, LAMP reaction mixtures and DNA binding fluorophores like SYBR Green I are added prior to amplification of nucleic acid and identification of bacterial species, respectively. Lastly, the sample containing bacterial DNA is added and analyzed using a UV transilluminator (Fig. 5A). The interaction of biotin with streptavidin promotes the attachment of primers to the membrane, thus improving its stability, specificity, and LOD of MRSA (Fig. 5B)126. As an alternative to SYBR Green I, metal complex-based probes have been investigated as fluorophores to reduce the possibility of false positives127. These metallic probes comprising phenazine, phenanthroline, and dipyridophenazine serve as a photoswitch. Upon the intercalation of the metal complex with DNA, the nitrogen atoms of phenazine get shielded, thus causing a change in the microenvironment. Ultimately, this interaction generates a high fluorescence which is used for the detection of genes present in AMR bacterial species. Zhou et al. developed paper-based chips with metal complexes for the simultaneous detection of multiple AMR genes127. The chips are made of multiple Whatman filter paper discs containing metal complexes as LAMP reactants. (Fig. 5C). Antibiotic-resistant genes (mecA and ermC coding for methicillin resistance and lincosamide-streptogramin resistance, respectively) were loaded on different spots of the paper chip, and the interaction between the genes and LAMP reactants was measured as fluorescence signals. As observed in Fig. 5D, the spots 2, 3, 4 containing 16S rDNA, ermC, and mecA, respectively, showed high fluorescence signals, whereas spot 1 (no template control) completely lacked fluorescence (Fig. 5D). This approach improved sensitivity and enabled multiplex detection of several AMR species. At the same time, in comparison to other fluorescence approaches for AMR sensing, it required relatively higher sample amount and a longer time for identification (Table 2).

A Paper-based LAMP device reaction pad comprising LAMP reactants, florescent dye, and sample (MRSA) of interest. Reprinted (adapted) with permission from ref. 126. Copyright {2021} American Chemical Society. B Fluorescence signal detecting MRSA positive sample using paper-based LAMP device. Reprinted (adapted) with permission from ref. 126. Copyright {2021} American Chemical Society. C Paper-based chips comprising five layers—magnetic plate, magnetic ports, filter paper, laminate, and magnetic plates. The isothermal amplification causing interaction between metal complex and antibiotic-resistant genes generates a fluorescence signal. Reprinted (adapted) with permission from ref. 127. Copyright {2018} American Chemical Society. D Multiplex detection of antibiotic-resistant genes in spot 2 (16S rDNA), spot 3 (ermC), spot 4 (mecA), and spot 1 as negative control. Reprinted (adapted) with permission from ref. 127. Copyright {2018} American Chemical Society.
Laliwala et al. have developed paper microzone plates immobilized with fluorescent ratiometric sensor array for the detection of individual bacterial species, Gram status128, as well as AMR S. aureus129. Paper microzone plates containing hydrophilic wells surrounded by hydrophobic barriers were fabricated using photolithography. The ratiometric sensor array consisted of four fluorescent dyes (derivatives of 3-hydroxyflavone) that portioned into the bacterial cell envelope using hydrophobic or π–π interactions. These interactions generated unique signals that were processed using pattern recognition algorithms. Machine learning algorithms, including linear discriminant analysis and neural network, differentiated individual strains of methicillin- and vancomycin-resistant S. aureus from their non-resistant strains. Additionally, this sensing approach differentiated clinical isolates as well as biofilms associated with MRSA and MSSA with an accuracy >80%129. This multiplex sensing approach overall aims to improve the robustness and cost of sensing AMR S. aureus strains.
Surface-enhanced Raman spectroscopy
Surface-enhanced Raman spectroscopy (SERS) was first discovered in 1974130. It is a spectroscopic technique that is based on the Raman effect – the inelastic scattering of light. In SERS, nanostructured surfaces or substrates are used to enhance the Raman scattering, thus enabling the identification of molecules present at low concentrations. Typically, SERS substrates comprise metals such as silver or gold, which exhibit a unique SPR effect. The SPR effect occurs due to the resonance between the electrons from the metal surface and the analyte of interest131. Thus far, to our knowledge, there is only one report utilizing paper-based SERS for the identification of AMR bacterial species132. In this approach, silver nanoparticles immobilized on paper were used as a SERS substrate (Fig. 6). A library of SERS reporter molecules was generated to identify antibiotic-modifying enzymes like β-lactamases. These reporter molecules have a SERS marker—a sulfur group that directly binds to the metal. The library of molecules is mixed with bacterial species that are potentially resistant to antibiotics and placed on a paper SERS sensor. Their interaction generates a barcode signal, enabling the detection of a specific analyte132. Therefore, the use of paper-based SERS provides a low-cost, portable, and easy-to-use technique for the identification of AMR species.
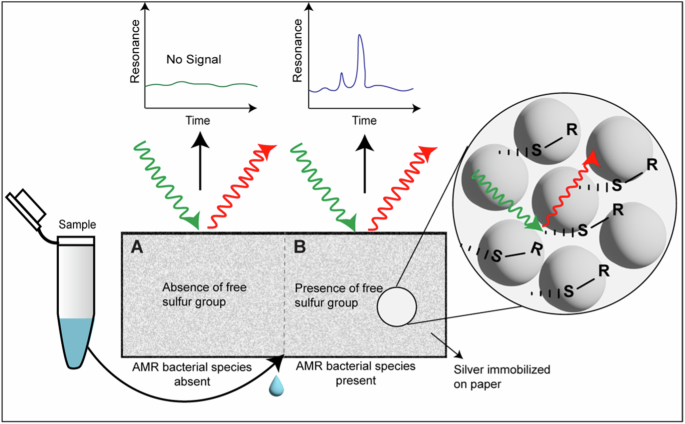
The absence of AMR bacterial species will generate a Raman spectrum with no signal (A). In the presence of AMR, bacterial species having β-lactamase activity will hydrolyze the recognition element to generate a sulfur-containing SERS reporter molecule (B). The interaction between the sulfur group and silver generates a unique Raman spectrum, thus enabling β-lactamase activity and the presence of AMR bacterial species. Adapted from ref. 132. Copyright {2020} by Elsevier. Reprinted with permission.
Nanoparticles in AMR detection
Nanoparticles can have unique properties and functionalities, making them an important instrument for sensing pathogenic bacteria species. Properties, including size, surface area, morphology, and charge, make them advantageous for targeting bacterial species. Due to the inherent spectral properties, functionalization ability, and ease in encapsulation of fluorescent probes, various types of nanoparticles have been used as receptors for colorimetry, fluorimetry, or SERS detection. Polymeric, gold, and silver nanoparticles, and magnetic nanoparticles have been investigated for their potential in sensing pathogenic bacteria133.
Various nanosized systems, including nanoparticles have been developed for the identification of AMR bacterial species134. For example, paper-based band-aids have been developed as a POCT for the detection of drug-sensitive and drug-resistant E. coli135. These band-aids were fabricated using a metal-organic framework coated with chitosan and chromogenic substrates like bromothymol blue and nitrocefin. The metal-organic framework was immobilized on cellulose paper to identify bacterial species as well as the presence of drug resistance. The presence of bacterial species changed the pH of the band-aid, which converted bromothymol blue from green to yellow. Next, drug resistance was determined by the change in the band-aid color from green to red. This occurred due to the release of a drug-inactivating enzyme, β-lactamase, that interacted with another chromogenic substrate, nitrocefin. This technique demonstrated a low LOD with 104 CFU/mL, 2–4 hours of analysis time, and high specificity. Another strategy for the identification of AMR bacteria includes the use of beta-iodometric test136. In this technique, the Whatman paper is covered with chitosan nanoparticles coated with a starch-iodine indicator. Subsequently, antibiotics (penicillins, cephalosporins, and carbapenems) responsive to β-lactamases were layered on the test strip. Bacteria resistant to antibiotics release β-lactamases that hydrolyze antibiotics to form penicilloic acids. In the presence of penicilloic acids, the starch-iodine complex (blue color) decolorizes. Despite this technique being relatively less sensitive, it significantly improved the analysis time for the identification of AMR bacterial species. To improve the specificity of AMR identification, paper-based multiplex analytical devices have been fabricated. In this technique, multiplex detection of enzymes and toxins has been developed for the identification of Clostridioides difficile137. This approach utilizes gold nanoparticles to enhance the signal intensity and improve the sensitivity.
Conclusions and outlook
The development of AMR in bacterial species has been on the rise for the past 50 years. Despite the advancements in the diagnostic tools currently available for the identification of AMR species, they are time consuming and tend to show false positive results. Furthermore, these tools lack user-friendliness and heavily rely on expensive instruments and trained personnel. To overcome these limitations, POCTs that meet the WHO’s REASSURED criteria are the subject of extensive research. POCTs that meet these required criteria enable the translation of diagnostic tools from bench to bedside, thus improving the outcome at the consumer level. Paper has multiple advantages where the morphology of the cellulose matrix allows the immobilization of recognition elements to improve the accuracy, sensitivity, and selectivity of the platform. Therefore, paper-based sensors are also actively investigated as POCTs. Paper can be designed as a sensing tool that is compatible with different detection techniques. Paper is lightweight, portable, and inexpensive, which significantly reduces the cost of paper-based devices.
In this review, we highlight the paper-based platforms that were developed for the sensing of AMR bacterial species. Paper-based sensors for AMR species identification use optical detection techniques like colorimetry, fluorimetry, and surface-enhanced Raman spectroscopy. A wide range of targets have been investigated. Using whole cells reduces the analysis time due to the absence of additional preparation steps, however, using other targets like enzymes and nucleic acids improves the accuracy and requires very low sample amount. Using colorimetric detection and targeting nucleic acids is a rapid, reliable, cost-efficient, and sensitive method for the identification of AMR species with the potential to translate into a diagnostic tool for testing clinical samples. Fluorometric detection using LAMP offers an extremely sensitive approach to the identification of AMR species. The use of a paper platform not only shortens the analysis time but also makes the sensor easier to use, more robust, and reliable. Despite having similar limits of detection and analysis time using colorimetric and fluorometric detection techniques, fluorometric detection enables the simultaneous detection of multiple species. Using nanoparticles to identify antibiotic-resistant bacterial species, thus far, only colorimetric and SERS detection have been employed. These techniques enable the multiplex detection of resistant species by interacting with enzymes, nucleic acids, and other proteins, thus improving the sensitivity.
At the same time, paper-based diagnostic methods face a number of challenges before they can be introduced into clinical practice as reliable diagnostic tools. While the sensitivity of antibody-based sensors is often quite high, the use of antibodies implies relatively high production costs and demanding requirements for handling. In some cases, to reach a diagnostic decision, some quantification of the analyte is required. While visual (particularly colorimetric) detection is the simplest and most user-friendly, it can only be reliably used for qualitative (or, at most, semi-quantitative) assessment. Besides this, reproducibility and batch-to-batch consistency in production may also appear as potential challenges.
To overcome these and other hurdles, further progress will follow in the integration of available handheld technology, such as smartphones, into the quantification process. Novel sensing elements will continue to be developed that will offer superior sensitivity, and aptamer-based sensors hold significant promise in this area. In addition, multiplex detection is gaining increasing attention, which will help increasing reliability and functionality of future paper-based sensors. On-board sample preparation (or collection) will likely see increased attention in the future, with dynamic advances in research featuring soft microneedle techniques.
Though limited research has been conducted in this field, it holds a significant promise for the development of future POCT. Paper-based sensors can be designed to accurately identify species with low sample volume and without the need of sophisticated instruments, shows its potential as a diagnostic tool, especially in limited-resource settings.



Responses