Developing a named entity framework for thyroid cancer staging and risk level classification using large language models
Introduction
Thyroid cancer is the most prevalent endocrine cancer and the 7th most common cancer type across the globe1,2. Although the mortality rate of thyroid cancer is relatively low (0.44 per 100,000) compared to other cancers, its incidence has surged by 313% over the past 40 years, reaching 9.1 per 100,000 worldwide in 20222,3. Differentiated thyroid cancer, predominantly papillary (~84%) and follicular (~4%) thyroid cancer, is the most common pathological subtype accounting for over 90% of all thyroid cancer cases3,4.
The 8th edition of the American Joint Committee on Cancer (AJCC)/Tumour-Node-Metastasis (TNM) staging system and the American Thyroid Association (ATA) risk stratification system are frequently used by clinicians who manage thyroid cancer5,6,7. The 8th edition of AJCC/TNM staging system is optimised to predict the survival of patients with thyroid cancer. It retains the basic anatomic pathology T-N-M staging approach and stratifies patients by the age of 556. Meanwhile, the ATA risk stratification system predicts the risks of disease recurrence or relapse and categorises patients into three risk groups (i.e., low, intermediate, and high) based on the characteristics of thyroid cancer such as tumour size, presence of aggressive cancer variants, extra-thyroidal extension, vascular invasion, lymph node involvement, etc5.
Information that determines cancer staging and ATA risks of patients with thyroid cancer is usually stored in lengthy unstructured or semi-structured clinical notes. As a result, clinicians take considerable time to manually retrieve critical information from multiple clinical notes to make decisions, potentially hindering prompt treatment provision and compromising the quality of patient care. Moreover, extraction of clinical information for research purposes from a large amount of unstructured data could be labour intensive. With the recent advancement in artificial intelligence (AI), Large Language Models (LLMs) demonstrated their capabilities to efficiently extract data from clinical notes8,9,10. LLMs accomplished various tasks of zero-shot learning, information extraction, and text summarisation11,12. Furthermore, providing LLMs with specialised, domain-specific datasets would further endow the LLMs with domain-specific knowledge and potentially reduce model biases13. In this regard, a framework that leverages the power of LLMs could in principle reduce the time and effort required for manual review, thereby helping clinicians optimise treatment decisions in a timely matter and improve patient outcomes. Moreover, an efficient and accurate tool could aid the conversion of huge unstructured clinical data into well-formatted structured databases, hence accelerating research in various medical fields.
There are three existing frameworks with different LLMs and prompting strategies developed using pathology reports for patients with thyroid cancer. A rule-based classification was built to extract stage-related information from full or semi-structured free-text clinical documents and transform the data to a standardised common data dictionary, indicating that these tools could help with data standardisation for observational research14. Another rule-based pipeline, ThyroPath, was developed for information extraction and tested on structured reports for risk classification of papillary thyroid cancers on a scale modified from the ATA risk categories10. Lee et al. developed a tool using a localised FastChat-T5 3B LLM to extract information from surgical pathology reports through a medical question answering (MQA) approach. The tool achieved an overall accuracy of 90% and significantly reduced the time spent on report reviewing compared with human15.
To date, no existing named entity (NE) framework has been specifically developed for extracting information from semi-structured or unstructured clinical notes to assess both the AJCC/TNM cancer staging and ATA risk categories in both types of well-differentiated thyroid cancers (papillary and follicular thyroid cancer). In this study, we addressed this gap by (1) developing an NE framework which consists of annotation guidelines, ground truth labels, and prompt and evaluation codes, and (2) examining different LLM strategies based on the developed NE framework. This framework enables extraction of local cancer-related information from semi-structured free-text clinical notes, followed by an ensemble of offline LLMs, thereby providing a secure and accurate tool for cancer staging and risk classification in patients with well-differentiated thyroid cancer.
Methods
Ethical considerations
This study received approval from the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW 24-319). Informed consent from patients was not required because of using the pseudo clinical notes and open-source clinical note dataset.
Data source
Pathology reports and clinical characteristics of all 507 patients with thyroid cancer were obtained from a public dataset—The Cancer Genome Atlas Thyroid Cancer (TCGA-THCA) programme. TCGA is a landmark cancer genomics programme. It molecularly characterised over 20,000 primary cancer and matched normal samples from 11,000 patients spanning 33 cancer types16. TCGA-THCA is a subproject that focuses on thyroid cancer under the TCGA programme. TCGA data are available without restrictions on their use in publications or presentations17, and they can be downloaded from National Cancer Institute (NCI) Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov).
Pathology reports sourced from TCGA-THCA programme are semi-structured data with subtitles and contain necessary information for cancer staging and ATA risk classification, including tumour sizes, number of lymph node resected and involved, histological subtypes of thyroid cancer, extrathyroidal extension status, presence of capsular and vascular invasion, margin involvement, distant metastasis, mutation status, etc. The TCGA-THCA programme also provided patients’ age and cancer stages using AJCC editions 4, 5, 6 and 7.
Among all the 507 patients from TCGA-THCA, 351 patients who were staged with the 7th edition of AJCC were included for further screening. Then, 12 patients were removed because their pathology reports did not provide sufficient information for cancer staging and/or ATA risk classification upon manual review (n = 11) or age below 18 (n = 1). A total of 339 patients remained, and they were split into two groups – 50 in the development set and 289 in the validation set. The ground truth for both 8th edition of AJCC cancer staging and ATA risk categories was generated for all 339 patients. Among these patients, 286 were classified as stage I, 47 as stage II, 4 as stage III, and 2 as stage IVB according to 8th Edition of AJCC system. The stage distribution aligned with large population-based epidemiological data (e.g. cancer registries), where stage I and II accounted for over 90% of all new thyroid cases18,19,20. In terms of ATA risk categories, 143, 122, and 74 were classified as ‘low’, ‘intermediate’, and ‘high’ risks, respectively.
For the NE framework development, a representative sample of 50 pathology reports with sufficient patients within each stage was included. Although a larger sample size is generally preferable, there is no theoretical minimal sample size for the NE framework development. As such, in order to reserve most of the samples for further validation, we selected 50 patients for training by oversampling patients with stage III or above and maintaining the percentage of patients with stage I and II at around 90%. The resulting training data comprised 31, 15, 2, and 2 patients with stage I, II, III, and IV, respectively.
The remaining 289 TCGA reports were used for further validation on the 8th edition of AJCC cancer staging and ATA risk classification. Figure 1 shows the flowchart for the compilation of our training and validation data.
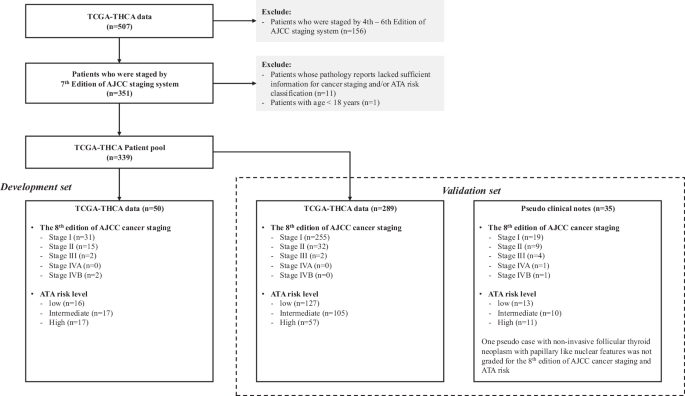
Flowchart depicting patient selection and the data source used as development set and validation set. Cancer stages and ATA risks of all TCGA-THCA patients and pseudo cases were verified by endocrine surgeons. A pseudo case of non-invasive follicular thyroid neoplasm with papillary like nuclear features is not grade with AJCC staging and ATA risk.
NE framework development
The NE framework included annotation guidelines, independent annotation by two annotators, ground truth labelling by clinicians, and prompts with various strategies for local LLMs to extract cancer-related disease information from clinical notes, and classification rules for classifying cancer staging and risk level using the LLM outputs.
For annotation, our team, consisting of endocrine surgeons (M.F. and Y.L.), clinical oncologist (V.L.), and an expert in developing and implementing AI models in clinical settings (Z.W.), co-developed the annotation guidelines (Supplementary Note 1). Necessary information extracted for classifying cancer stage according to the 8th edition of AJCC cancer staging system7 and ATA risk category5 was detailed in the annotation guidelines. In total, there were 29 named entities, 1 relation, and 1 attribute. Using the text annotation tool Brat (http://brat.nlplab.org/)21, our annotators (T.W. and W.Y.C.) who are experienced medical researchers performed annotation independently. If there was any disagreement, two expert annotators (M.F. and Y.L.) would discuss with the annotators and resolve it with their expertise. A F1-score (equivalent to kappa agreement rate) of over 80% was assessed upon the completion of human annotation22,23. Annotated data for each case were stored in .ann files. The step-by-step annotation process and examples were provided in Supplementary Note 2.
To establish the ground truth, the semi-structured clinical notes of all 339 TCGA cases (50 for development and 289 for validation) were manually reviewed, and each assigned with an 8th edition of AJCC cancer staging and ATA risk category. The ground truth of each case was verified and validated by two endocrine surgeons (M.F. and Y.L.) (Supplementary Data 1). The age of patients was obtained from cross-referencing with the TCGA dataset using unique patient study identifiers.
To perform information extraction, we formulated an inference prompt that directed the LLMs to identify all the named entities of interest from a clinical note. This inference prompt comprised an instructional segment and a contextual segment constituted by the semi-structured clinical note. A specific instruction was designed for each NE in the inference prompt, such as determining whether a unit should be included in the output, so that the model was more likely to extract pertinent information. The Python package ‘Langchain’ was adopted to create the inference prompt templates which would subsequently be run in LLMs24. To ensure that the LLMs would generate the same output given the same data, we set the temperature value to 025,26. Besides, we adopted JSON parser in ‘Langchain’27,28 which allows us to only capture the JSON outputs and ignore the irrelevant text generated by the LLMs. Eight prompting approaches were developed and applied to LLMs. First, zero-shot prompting was used as the baseline performance of extracting disease information. The second approach was zero-shot COT prompting, encouraging LLMs to explain their reasoning process in a stepwise approach, without providing any examples29,30. Few-shot prompting with annotation data of 50 TCGA pathology reports (the development set as mentioned above) was used for the third to fifth approaches. The third approach was providing all the annotated information in the corresponding entity in the inference prompt. The fourth approach was adding the annotated information in the corresponding entity but reducing the amount of information by eliminating those with similar meaning. Using ‘CompResectPath (Completeness of resection from a pathologist perspective)’ as an illustration, both wordings ‘free of tumor’ and ‘FREE’ were provided as examples of clear surgical margin in the third approach, but only ‘free of tumor’ was kept in the fourth approach. The fifth approach was only adding the annotated information about the site of gross extrathyroidal extension, level and site of lymph node, and histologic subtype of thyroid cancer because such information was frequently being overlooked by the zero-shot algorithms. The sixth to eighth approaches combined COT prompting (i.e., the second approach) with various extent of annotated data (i.e., the third to fifth approaches). A step-by-step process of designing few-shot prompts using annotated data were provided in Supplementary Note 3, and an example detailing the inputs and outputs of LLMs using different prompts was presented in Supplementary Note 4.
We prepared a Microsoft Excel® template, which contained our self-developed algorithms (i.e. Formula and VBA coding), to store and clean the LLM raw outputs, and then to classify the AJCC 8th edition cancer staging and the ATA risk category of each patient (Supplementary Data 1). The cleaning steps involved standardising length unit (e.g., converting tumour size from millimetres to centimetres) and removing unnecessary information from the raw outputs, including extra symbols, words, and irrelevant information in various entities (e.g., lymph node information in “distance metastasis”). The embedded Excel algorithms then used these processed data to perform cancer staging and risk classification. The 8th edition AJCC cancer staging system31 is one of the most widely used staging systems for thyroid cancer. It categorises each patient according to tumour (e.g., tumour dimensions, margins, involvement of adjacent structures), lymph node (number and location), and distant metastases status. It is used to predict disease survival. The ATA risk stratification system is another commonly used system to predict disease recurrence and guides subsequent adjuvant treatment with thyroxine suppression and/or radioactive iodine. It categorises each patient into one of the three risk groups (‘low’, ‘intermediate’, and ‘high’) based on a wide range of tumour features, such as tumour size, extra-thyroidal extension, aggressive tumour variants, margins, lymph node involvement, etc5. A NE framework together with a classifier or LLM strategy addressing the above two systems would be the most relevant to clinical practice as they are widely adopted worldwide.
LLM strategies
We selected four offline LLMs for information extraction from semi-structured clinical notes in this study: Mistral-7B-Instruct-v0.3 (Mistral AI), Gemma-2-9B-Instruct (Google), Llama 3.1-8B-Instruct (Meta), and Qwen2.5-7B-Instruct (Alibaba). We chose these LLMs because of their state-of-the-art technology, openness, and lightweight nature which minimised the requirement of computational power and storage32,33. Most importantly, all these LLMs supported local deployment, enabling offline prompting and preserving patient privacy when real clinical notes are used. Ollama, an open-source software platform, was used to operate the local LLMs in this study34.
In addition, majority-voting strategy was adopted to evaluate the ensemble performance of LLMs and approaches35. Two majority-voting approaches were conducted, namely (1) at outcome level and (2) at factor level. For outcome level, we used majority-voting to generate the ensemble outputs for ATA risk and AJCC 8th edition cancer staging from the classification results on the outputs given by all model-prompt combinations. For factor level, we applied majority voting on each relevant factor that was used for classification of ATA risk and AJCC 8th edition cancer staging. As a result, one set of ensemble factors were obtained. The ensemble factors were then used to generate the ensemble cancer staging and risk classification.
Evaluation of the LLMs with framework development set
To assess the performance of the LLMs in framework development, we compared the LLM-extracted 8th edition of AJCC cancer staging and ATA risk categories against the ground truth of the representative sample of 50 TCGA reports as the framework development set. The F1-score, a commonly used performance metric for extraction tasks, was applied to provide a balanced assessment of model precision and recall. To adjust for differences in sample size among each risk level and staging category, weighted average of F1-score, precision and recall were calculated. A higher score indicates a better performance of the LLMs that meet our expected standards.
Evaluation and validation of LLMs with validation set
Two data sources were used as the validation set for further evaluating and validating the NE framework. First, 289 pathology reports from the TCGA-THCA programme (as mentioned above) were used for validation on the AJCC 8th edition stage and ATA risk categories. Second, clinical notes of 35 pseudo cases, which were created and labelled with ground truth by two endocrine surgeons (M.F. and Y.L.), were used for validation. Unlike TCGA-THCA dataset that documented all clinical features in pathology reports, each pseudo case had one operation record, which was the main source of surgical margin status and presence of gross extrathyroidal extension, and at least one corresponding histopathology report, where most other clinical features can be found at. In local clinical practice, the clinicians would refer to both types of clinical notes to make decisions. There were some differences in the formats or expression of entities between TCGA clinical notes and local clinical notes. For example, majority of TCGA clinical notes only included the site of extrathyroidal extension without specifying whether it being gross or microscopic, whereas local clinical notes would provide information on the extent of extrathyroidal extension (gross or microscopic). The pseudo clinical notes were created to resemble the format and contents of semi-structured clinical notes in Hong Kong, as we intend to also apply our established NE framework and LLM strategies to local clinical practice, while real clinical notes were currently inaccessible for this study due to data privacy concerns. Furthermore, there were no patients with stage IVA or non-invasive follicular thyroid neoplasm with papillary like nuclear features in the TCGA-THCA dataset, and thus the pseudo cases supplemented the TCGA-THCA dataset’s limitations. The details on the pseudo clinical notes creation process are available in Supplementary Note 5.
The flow of the data extraction using LLMs and classifying ATA risk and AJCC staging was depicted in Fig. 2 and an example was used in Supplementary Note 4.
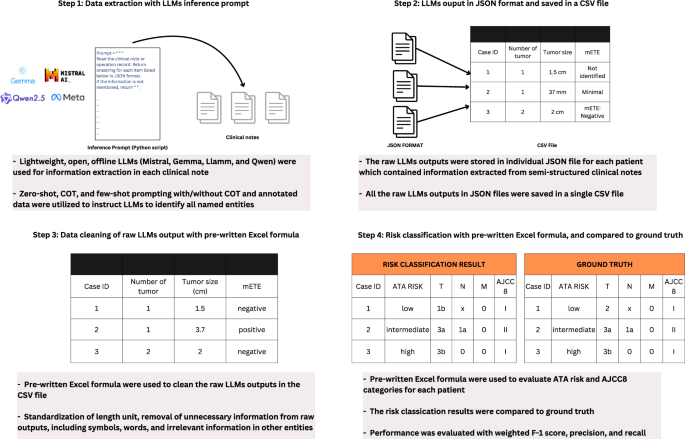
Schematic diagram depicting the flow of data extraction using LLMs and the utilization of self-developed Microsoft Excel template for data cleaning and classification.
Results
Patient characteristics of the development set
The characteristics of the 50 selected TCGA-THCA patients for the NE framework development were displayed in Table 1. The mean age of the patients were 54.3 (SD 13.9) years. The majority of the patients were female, white. Both papillary and follicular carcinomas were included, at proportions consistent with real-world observations. After the validation by the two endocrine surgeons (M.F., and Y.L.), there were 31, 15, 2, and 2 patients staged as I, II, III, and IVB based on the 8th edition of the AJCC system respectively. Each of the three ATA risk category (low, intermediate, high) has around one-third of patients.
Results on the LLMs performance with development set
The kappa agreement rate between the two annotators was 84.3%, ensuring acceptable inter-rate reliability. All LLMs with few-shot prompting attained F1-scores of 90.3–100.0% for the 8th edition of AJCC cancer staging, and 88.0–100.0% for ATA risk classification (Fig. 3 and Supplementary Table 1). The F1-scores on ATA risk and AJCC overall staging were 100.0% and 94.1% for the ensemble classifier, respectively (Fig. 4 and Supplementary Table 1). Of note, all zero-shot and few-shot models achieved an F1-score of 100.0% for the M stage (Supplementary Table 2). However, while most models demonstrated an F1-score of approximately 90.0% for the T and N stages, the F1-score of Mistral-7B-Instruct-v0.3 using COT and few-shot prompting with non-repeated annotated data was below 80.0% for the N stage (Supplementary Table 2). Gemma-2-9B-Instruct using few-shot prompting with part of annotated data seemed to be the best model, reaching F1-scores of 100.0% for both ATA risk and 8th edition of AJCC cancer staging (Fig. 3 and Supplementary Table 1).
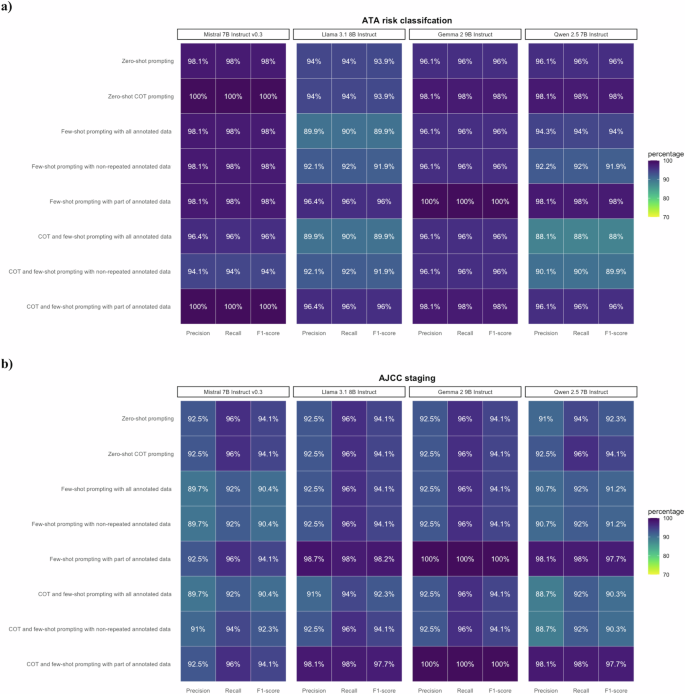
LLMs with various prompting strategies attained satisfactory performance in NE framework development. a Performance on ATA risk classification with F1-scores 88.0–100.0%. b Performance on AJCC staging with F1-scores of 90.3–100.0%.
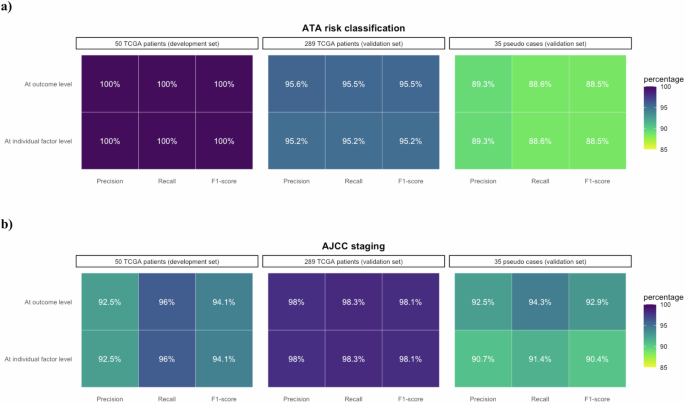
Ensemble classifiers attained satisfactory performance on the two datasets. a Performance on ATA risk classification with F1-scores at least 88.5%. b Performance on AJCC staging with F1-scores of at least 90.4%.
Validation of LLMs with validation set
The characteristics of 289 TCGA-THCA patients and 35 pseudo cases within the validation set are presented in Table 1. For the 289 TCGA-THCA patients, the F1-scores of all four LLMs ranged from 88.5 to 96.5% for ATA risk classification and 94.2–99.7% for AJCC cancer staging, and the ensemble classifiers achieved F1-scores of 95.2–95.5% and 98.1% in ATA risk classification and AJCC cancer staging, respectively (Figs. 4, 5, and Supplementary Table 3). Most prompting strategies attained an F1-score of 100.0% for the M stage, and all strategies achieved an F1-score of over 90.0% for the T stage (Supplementary Table 4). While the F1-scores of Mistral-7B-Instruct-v0.3 for the N stage were 80.1–86.5%, those of other models were all above 90.0% (Supplementary Table 4).
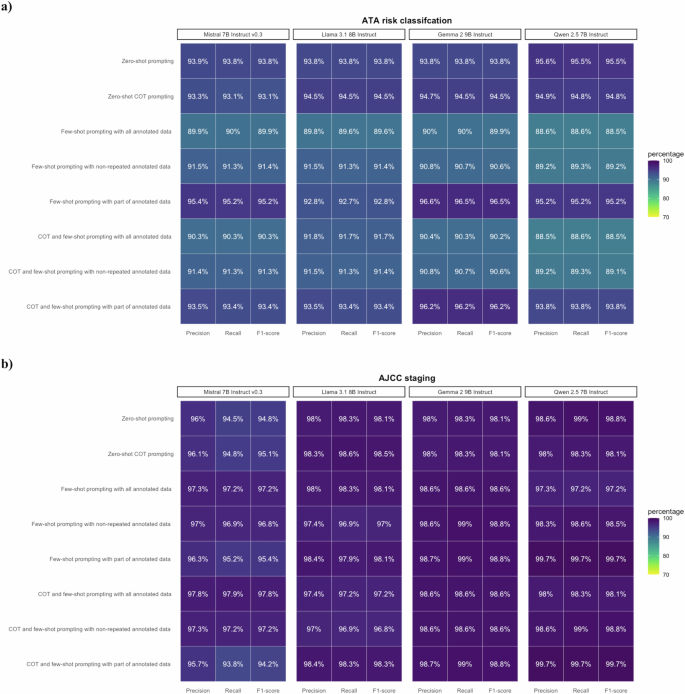
LLMs with various prompting strategies attained satisfactory performance in 289 TCGA pathology reports for validation. a Performance on ATA risk classification. with F1-scores 88.5–96.5%. b Performance on AJCC staging with F1-scores 94.2–99.7%.
For the 35 pseudo cases, the F1-scores for the ensemble classifier on ATA risk and AJCC staging were 88.5% and 90.4–92.9%, respectively. (Fig. 4 and Supplementary Table 5). Mistral-7B-Instruct-v0.3 outperformed the other models in ATA risk classification (with highest F1-score of 94.3%), while Llama-3.1-8B-Instruct had the best performance in AJCC cancer staging (with highest F1-score of 97.5%) (Fig. 6 and Supplementary Table 5). Except Llama-3.1-8B-Instruct using COT and few-shot prompting with part of annotated data had an F1-score of 96.7%, all other prompting strategies achieved F1-scores of 100.0% for the M stage (Supplementary Table 6). The F1-scores for the T and N stages ranged from 60.1 to 81.9% and 71.9 to 97.2%, respectively (Supplementary Table 6).
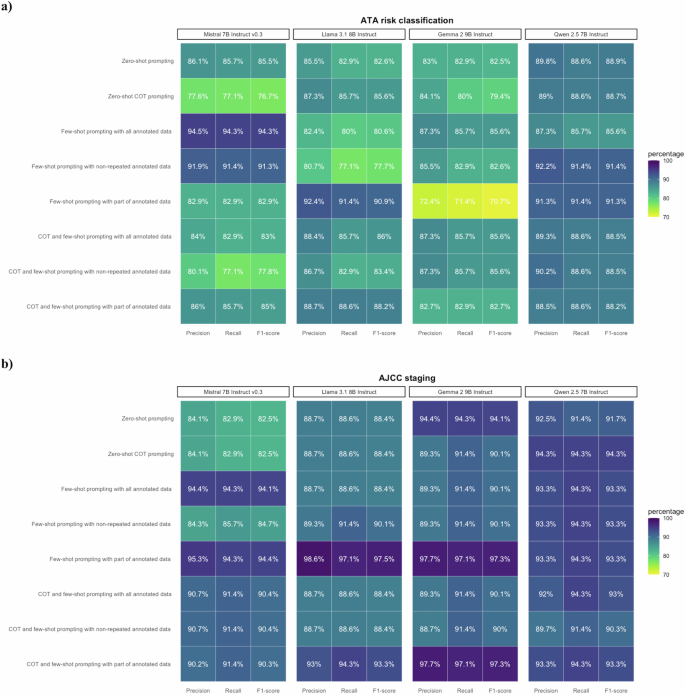
The performance of LLMs various in different approach and in individual LLM in the 35 pseudo cases for validation. a Performance on ATA risk classification. Mistral-7B-Instruct-v0.3 outperformed other LLMs with F1-score of 94.3%. b Performance on AJCC staging. Llama-3.1-8B-Instruct outperformed other LLMs with F1-score of 97.5%.
Misclassification investigation
The confusion matrices of AJCC cancer staging and ATA risk classification were created for the misclassification investigation (Supplementary Tables 7–12). Within the framework development set, Mistral-7B-Instruct-v0.3 had better performance in classifying patients with ‘intermediate’ and ‘high’ ATA risks, as most prompting strategies employed in the other three LLMs reported misclassifications within those two ATA risk categories. However, Mistral-7B-Instruct-v0.3 was found to have more misclassifications when classifying patients with ‘low’ risks (Supplementary Table 7). All four LLMs had misclassifications with ‘Stage III’ of the 8th edition of AJCC cancer staging (Supplementary Table 8). For the validation set, most LLMs reported misclassifications across all three ATA risk categories and ‘Stage III’ among the 289 TCGA patients (Supplementary Table 9–10). The LLMs reported misclassification among cases with ‘intermediate’ and ‘high’ risk categories of ATA for the 35 pseudo cases. Of note, only few models successfully classified patients with Stage IVA. This may be due to the absence of stage IVA patients in the initial NE framework development taken from TCGA (Supplementary Table 11–12).
We further investigated the reasons behind the misclassification and noticed that LLMs tended to generate incorrect answers when identifying the extent of extrathyroidal extension, the largest size and number of the involved lymph nodes, aggressive variants of papillary thyroid cancer (histologic subtype), the presence of vascular invasion, and the completeness of surgical margin (Supplementary Tables 13–15). Supplementary Figures 1–3 visualise the number of errors leading to misclassification of AJCC cancer staging and ATA risk classification. All 4 LLMs had examples of misclassifying the entity “extrathyroidal extension”, leading to incorrect AJCC cancer staging. This is because, in many of the TCGA pathology reports, the entity ‘extrathyroidal extension’ was described in various and rather ambiguous ways, without explicit description on it being gross or microscopic (e.g. ‘extrathyroidal extension: invades: perithyroidal tissue’, ‘extrathyroidal extension: focally present’, ‘left with extrathyroidal extension’, ‘extrathyroidal extension: yes’, etc.). Since the extent of extrathyroidal extension (gross vs microscopic) was an important discriminant factor for the ATA risk categories (‘intermediate’ or ‘high’) and T stage (T2 vs T3 or above) of the 8th edition of AJCC cancer staging, errors in this entity would affect the model performance. In addition, each LLM failed to correctly capture the largest size and/or number of involved lymph nodes for up to 2 TCGA patients. Indeed, each pathology report may provide sizes of multiple items, such as specimens, both involved and uninvolved lymph nodes, and tumours. This complexity could potentially lead to confusion for LLMs extracting the largest dimensions of the involved lymph node. Moreover, the LLMs may be confused with the number of lymph node involved and the number of lymph node resected, and it may have difficulties in providing the total number of involved lymph nodes if this information was presented separately according to the anatomic positions without a summary total of all involved lymph nodes. While the accuracy in identifying the lymph nodes status and anatomical location was crucial for accurately assigning the N stage in AJCC cancer staging, the number and size of involved lymph nodes affected the results of ATA risk classification. Other common reasons that led to misclassification of ATA risk categories included: with certain prompting strategies, some LLMs failed to extract the aggressive histological variants (e.g., Mistral-7B-Instract-v0.3 using COT and few-shot prompting with non-repeated annotated data used in the development set, all 4 LLMs used in the 289 TCGA validation set, etc), presence of vascular invasion (e.g., all 4 LLMs used in the 289 TCGA patients and Qwen2-7B-Instruct using zero-shot prompting used in the 35 pseudo cases from the validation set), and/or misjudged the surgical margin status (e.g., Llama-3.1-8B-Instruct used in the development set and 289 TCGA patients from the validation set, etc).
Discussion
Our works have developed an NE framework and LLM strategies with an adoption of an ensemble-like majority-voting strategy to automatically perform AJCC cancer staging and ATA risk classification for patients with thyroid cancer based on semi-structured free-text clinical notes. The LLMs achieved an accuracy exceeding 90.0% in cancer staging and ATA risk classification for the 50 TCGA-THCA patients, and achieved F1-scores of 94.1% and 100.0% respectively when employing an ensemble classifier. For further validation, the ensemble classifier achieved F1-scores exceeding 95.0% in both cancer staging and ATA risk classification for the 289 TCGA-THCA patients, and attained around 90.0% for the 35 pseudo cases.
The value of our study lies in its pioneering development of an NE framework, tailored for both AJCC cancer staging and ATA risk classifications, the two systems clinicians most frequently use to assess prognosis and determine subsequent adjuvant treatment and follow-up plans for patients with thyroid cancer. Furthermore, this study provided an example of annotation guidelines for future studies requiring human annotation. The use of lightweight LLMs that support local deployment could preserve the privacy of patients when the real clinical notes were used36. In addition, sharing human-annotated data on openly available TCGA pathology reports also enhanced data availability and encouraged research development in the fields of digital health. Although our framework was developed merely for clinical application in patients with thyroid cancer, this could potentially be extended to other cancer types or even other diseases. Beyond zero-shot prompting, COT prompting with or without varying degrees of annotated data was applied to enable reasoning capabilities within LLMs and investigate potential improvements in model performance.
The results of this study showed that the offline lightweight LLMs, namely Mistral-7B-Instruct, Gemma-2-9B-Instruct, Llama 3.1-8B-Instruct, and Qwen2-7B-Instruct, and the ensemble classifier are promising in solving practical extraction and classification tasks in an efficient and secure way12, suggesting the feasibility of adopting these tools in real clinical settings and research. The application of the LLMs can efficiently reduce the time clinicians spend on reviewing and cross-referring multiple lengthy clinical notes, thereby enhancing the efficiency of consultation and treatment, and improving patient care. However, the traceability into how the LLMs and ensemble classifiers generated the staging and ATA risk outputs was essential, as it may affect the clinicians’ decision to adopt our tool. Therefore, further improvements in highlighting the stage- or risk-related text used for risk classification in the original pathology reports would greatly enhance the LLM output’s traceability and transparency, vital for instilling clinician confidence and acceptance of its applicability. Conceivably, these features and interface are still under development but will be critical for implementation in real clinical settings.
In comparison to the methodologies and performance of other existing tools tailored for thyroid cancer patients, the ensemble classifier utilizing all LLMs exhibited a comparative accuracy of about 90% in both the cancer staging and ATA risk classification. Another rule-based pipeline, ThyroPath, had an 93% accuracy in risk classification based on the 2015 ATA guidelines using structured pathology reports. However, the task was not tested in non-structured pathology reports, characterised by free-text narratives and diversity in reporting variables, due to significant heterogeneity10. Our NE framework and LLM strategies, in contrast, were accurate in extracting information from free-text narratives and achieved satisfactory outcomes. Moreover, with the advances in machine learning techniques, there has been a continuing transition from traditional rule-based approaches to learning-based approaches, which do not require explicit manual coding of rules for each entity37,38. Consistent with Lee and colleagues, our study also illustrated that lightweighted localised LLMs could read and extract information from pathology reports within a short period of time15. Alternative prompt designs and strategies, particularly the multi-step extraction strategy, may boost the performance of LLMs39,40. However, when applied to the clinical notes in our study, such strategy yielded either similar or inferior performance compared to the prompting strategies that we have considered. This could be due to the absence of contextual information from other questions when extracting named entities individually, causing the LLMs to miss certain named entities. Unlike other existing framework or pipelines, ours incorporated annotated data and our results emphasised the value of integrating annotated data in maximising overall performance.
The major obstacles in developing our NE framework included the availability of ground truth and human-annotated data, prompt design, use of annotated data, and the inherent nature of LLMs. Firstly, our study highlighted the significant time and costs associated with ground truth generation—Endocrine surgeons (M.F. and Y.L.) manually reviewed information from clinical notes and classified the cancer staging and risk category for each patient, despite the availability in the current study. Similarly, the scarcity of high-quality, human-annotated data has been widely recognised as a challenge, with several literatures emphasising the expenses and difficulties in acquiring large-scale, well-annotated datasets41,42. Secondly, prompt design was a heuristic research process with many factors42. Numerous attempts were made to obtain a better model performance by modifying and testing the prompts. For example, we found that LLMs would return inconsistent formats of the T stage (e.g., ‘T3’, ‘pT1b’, and ‘4a’) or even hallucinate to produce unrelated outputs. To restrict the output formatting, we asked the LLMs to return desirable outputs by giving examples: ‘T1a’, ‘T1b’, ‘T2’, ‘T3a’, ‘T3b’, ‘T4a’, and ‘T4b’. Furthermore, we also applied COT and few-shot prompting by giving examples and outlining the reasoning process to enhance its problem-solving skills. Regarding the use of annotated data, we tested the performance of LLMs by using various extents of annotated data – we contrasted the outcomes by inputting all annotated data, annotated data without repetitive terms, and annotated data that were detected as challenging for LLMs to capture. Our study results suggested that the performance varied depending on the extent of annotated data employed. However, there was no definite conclusion that utilising a specific extent of annotated data would yield the optimal model accuracy. Moreover, the direction of changes in accuracy was inconsistent when using same extent of annotated data in different LLMs. In addition, the current study suggested that utilizing various prompting approaches, including different zero-shot and few-shot approaches, for the four LLMs did not produce remarkable differences in the accuracy of cancer staging and risk classification. This is consistent with other studies comparing different LLMs with different prompting approaches, and the performance would be context-specific and LLM-specific43,44. Therefore, it is imperative to conduct extensive experiments to acquire preferable results. Lastly, due to the ‘black box’ nature of the LLMs, the models lacked transparency on how the outputs were generated, and sometimes it was difficult to explain the reasons why LLMs produced an incorrect answer output even though the examples were given in prompts45,46. Due to its intrinsic stochastic nature, LLMs may respond to the same question with varying formats or even content when the question was repeatedly asked. Despite an overall model accuracy of over 90%, it is advisable that outputs generated by LLMs undergo human verification at this stage.
Several limitations of this study should also be acknowledged. Firstly, given that the TCGA-THCA programmes did not collect operation records and imaging reports for patients with thyroid cancer, we were unable to definitively distinguish whether the reported extrathyroidal extension was microscopic or gross in nature for some of the cases. Assumptions were made that the presence of extrathyroidal extension invading the skeletal muscles indicated the presence of gross extrathyroidal extension. Secondly, only 4 patients in the TCGA-THCA cohort were stage III or IVB, while none were stage IVA based on the 8th edition of AJCC cancer staging system. The small number of patients with stage III or above in the NE development set may affect the ability of the LLMs and classifier to devise outcomes in patients with advanced stages of cancer. Thirdly, we used the 8th edition of AJCC cancer staging system and 2015 ATA guideline to categorise patients in this study. However, the entities extracted under the NE framework and the classification rules may need to be updated when later versions of AJCC and ATA guidelines are released. Lastly, this study focused on LLMs with seven to nine billion parameters to balance the running time and computational power. The performance of larger LLMs, such as Mistral Large, Llama-3.1-70B and 405B, and Gemma-2-27B, was not evaluated.
In conclusion, this study initially constructed an NE framework, consisting of an annotation guideline, ground truth labels, LLM prompting, and evaluation codes; and secondly examined diverse LLM strategies, including four lightweight offline LLMs and ensemble-like majority voting strategies, to classify AJCC 8th edition thyroid cancer staging and ATA thyroid cancer risk category from semi-structured clinical notes. Our ensemble classifier optimised the efficiency and accuracy of cancer staging and ATA risk classification for well-differentiated thyroid cancer.

Responses