FeatureDock for protein-ligand docking guided by physicochemical feature-based local environment learning using transformer

Introduction
Drug discovery is a complicated, costly, and time-consuming task that typically takes pharmaceutical companies over a decade and billions of dollars to bring a novel drug to market1,2. The high-throughput screening (HTS) methods take considerable amount of time to evaluate a compound library that typically contains millions of compounds3. Considering the number of currently available compounds being ~108 in small molecule libraries such as PubChem4,5, ZINC6,7 and ChEMBL8, numerous in silico methods evolved to virtually screen potential compounds against the target to alleviate the heavy duty in HTS.
In the computer-aided and rational drug design, docking-based methods9,10,11,12 provide powerful tools for predicting the positions, orientations, and conformations of ligand binding. One of the critical principles of traditional docking methods is to design an accurate scoring function that can be applied to score and explore the optimal ligand binding poses. For example, DOCK13,14,15 and AutoDock16 used physics-based scoring functions consisting of terms for van der Waals (vdW) interactions, electrostatic energy, hydrogen bonding interactions, etc. These physics-based scoring functions consider inter- and intra-molecular chemical interactions, but they are often computationally expensive and suffer from the inaccurate descriptions of polarization, solvation and the entropy of protein and ligand17,18. Empirical scoring functions can address the above limitations of physics-based scoring functions by fitting parameters to the experimental binding affinities using pre-selected descriptors19,20. For example, AutoDock Vina21 utilizes a fully empirical scoring function, represented as vdW-like potential, hydrophobic interactions, conformational entropy, etc. Smina22 designed a user-friendly software interface to support custom scoring functions based on AutoDock Vina. Other derivatives of AutoDock Vina further improved the scoring function of AutoDock Vina for specific systems, e.g., AutoDock VinaXB23 that deals with halogen bonds and Vina-Carb24 that deals with carbohydrate systems.
The aforementioned docking methods employ genetic algorithms (GA)13,14,16 or Monte-Carlo (MC) annealing21,23,24,25,26,27, combined with gradient-based optimization, to sample and refine the ligand binding poses. More recent studies have integrated deep learning models to optimize ligand poses in one-stage28,29,30, significantly speeding up the docking process. For example, EquiBind30 and TankBind29 adopted E(3)-equivariant graph neural networks (E(3)-GNN) to effectively represent both the protein and ligand structures. They then utilized a graph matching network to predict the accurate rigid SE(3) transformation for the ligand docking. Additionally, a fine-tuning model was included to optimize torsion angles of rotatable bonds, enabling flexible docking. Unlike EquiBind and TankBind, which treated the docking task as a regression problem, DiffDock28 took a different approach by harnessing diffusion-based generative models. Specifically, it leveraged a denoising diffusion process to generate ligand conformations and binding poses from the initial ligand structure with randomized torsional angles and mean-removed 3D coordinates. DiffDock included a supervised model to classify whether the generated poses are close to the ground truth based on the model prediction called confidence score.
The scoring functions of docking methods are often inadequate in virtual screening, particularly when distinguishing strong binders from weak binders. This is because the scoring functions of docking methods often exhibit weak or no correlation between docking scores and binding affinities31,32. This is evidenced by the low Pearson correlation coefficients (Rc) between docking scores and binding affinities for several commonly used docking software evaluated on the CASF-2016 dataset32. For example, AutoDock Vina, GOLD33, MOE34 and Glide35,36 achieved Rc of 0.604, [0.416, 0.617], [0.405, 0.591] and [0.467, 0.513] respectively, where the range indicates that more than one scoring function are evaluated in the software. As a result, these docking scoring functions may result in high false positives when selecting true inhibitors by considering the highest-scored compounds as inhibitors from a compound library.
Recently, several machine-learning based scoring functions12,32,37 have engaged more comprehensive descriptors and more advanced neural network architectures to predict experimental binding affinities (e.g., Kd values), thus improving the scoring power. However, these machine learning methods were designed to re-score the docked pose, which could not apply to the scenario without 3D protein-ligand structures. For example, 3D Convolutional neural network (CNN) models like Atomic Convolutional Neural Networks38 and KDEEP39 treated protein-ligand complex as atom property tensors. Furthermore, distance-aware graph neural networks (GNN) like PotentialNet40 utilized the more efficient graph representation to encode Euclidean symmetries in structures.
In this study, we introduce a physicochemical feature-based deep-learning approach, FeatureDock, to score and pose small-molecular ligands binding to proteins. In FeatureDock, the potential binding pockets are firstly discretized into grid points, embedded using 3D-invariant FEATURE41 representations. These representations have been shown sufficiently descriptive to extract the local chemical environment for protein-RNA binding42. Then, the state-of-the-art Transformer encoder43 is adopted to predict probability density envelopes, formed by grid points in the binding pockets with high probabilities for ligands to occupy. The attention mechanism in Transformer further helps explain and visualize the importance of input features by extracting the attention weights. Using the probability density envelopes, we design a novel scoring function combined with a position optimization algorithm to screen and pose ligands in binding pockets. To demonstrate the performance of FeatureDock, we have applied it to select strong inhibitors from bioassay datasets8 for two different protein systems: an inactivated form of Cyclin-Dependent Kinase 2 (CDK2) and Angiotensin-converting enzyme (ACE). We show that our scoring function exhibits a superior ability to differentiate between strong and weak inhibitors compared to DiffDock28, Smina22 and AutoDock Vina21, as evaluated through the KL divergence and AUC evaluation. Notably, our model can accurately retrieve the binding poses of top-predicted ligands after the optimization process based on our scoring function. These binding poses closely (an average RMSD of 2.4 Å) approximate their native structures in cocrystal configurations, demonstrating a high level of proximity for CDK2.
Results
Model training and selection
We chose Transformer encoders in our FeatureDock, as it outperforms other neural network architectures, including FNN and ResNet (see details of these architecture in Methods Section “Neural network architectures”). The model performance was evaluated by cross-entropy loss, area under ROC (AUC), F1 Score and Matthews correlation coefficient (MCC) on the validation set. In detail, we performed a comprehensive hyperparameter tuning analysis for each architecture, exploring four different model sizes (Supplementary Table 3) and five learning rates, both with and without learning rate decays. As shown in Supplementary Fig. 15, Transformer encoder, with 500 K parameters and initial learning rate of 0.01 with plateau learning decay, delivers the best performance across all tested models. We further fixed the learning rate (0.01 with plateau learning decay) to solely evaluate the model performance among different model architectures. As shown in Fig. 1A and Supplementary Table 4, Transformer encoders consistently achieved the lowest loss across all tested models (FNN and ResNet), demonstrating the robustness of utilizing Transformer within our FeatureDock framework.
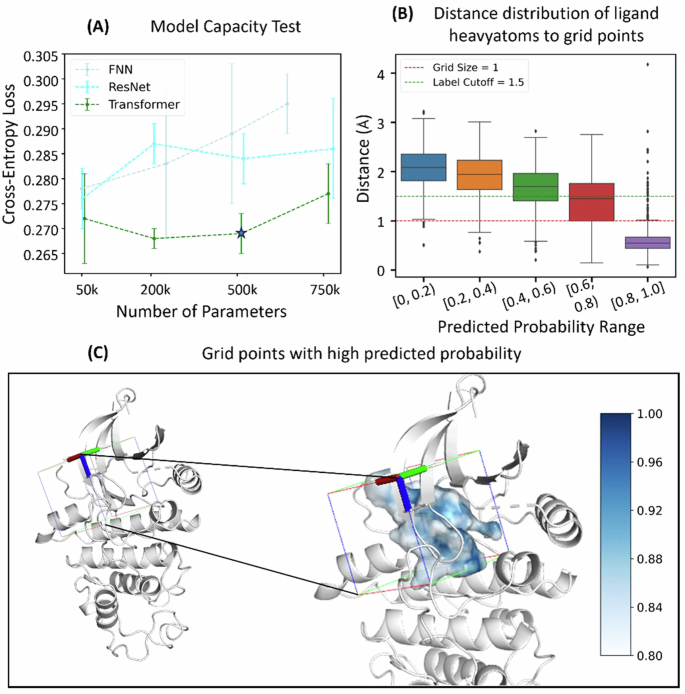
A Comparisons of cross-entropy loss of the validation set of three different architectures: FNN, ResNet, and Transformer. B Distance distributions of true ligand atoms to grid points. C Query box and the probability density envelope of 1B38 (inactive CDK2). The darker blue regions have higher probabilities.
To assess the applicability domain of our Transformer-based FeatureDock in terms of diverse chemical spaces as well as protein family in modern virtual screens, we first demonstrate that our model achieves consistent performance across a broad chemical space of ligand, validated by several evaluation metrics on ten representative functional groups that can cover an average of 82.30% heavy atoms of all ligands in the training dataset (see Supplementary Note 4 and Supplementary Fig. 16 for details). We further show that the fine-tuning on a specific protein family was not necessary in our framework. This is supported by the evidence that CDK2 leave-out model further optimized on kinase proteins did not show significant performance improvement over the general model (Supplementary Table 5) when testing on the CDK2 dataset.
Prediction of probability density envelopes for the ligand binding
We first demonstrate the performance of FeatureDock on CDK2 by training a model excluding CDK2 and its 90% homogeneous structures. FeatureDock can generate a probability density envelope by collecting the predicted binding probabilities of all grid points within the query pocket (see Supplementary Fig. 4 for the pocket information). Regions predicted with higher probability suggest a greater likelihood of being occupied by the ligand (Fig. 1C), making the probability density envelope a valuable tool for ligand design and ligand binding pose prediction. To assess the precision and robustness of our method’s predicted probability density envelope with regards to ligand occupancy, we analyzed 18 pre-aligned structures (Supplementary Fig. 4) of CDK2 (the inactive form) and CDK2/Cyclin (the active form). Figure 1B illustrates the distances between ligand heavy atoms and grid points associated with various predicted binding probability ranges. Notably, grid points with high predicted binding probabilities (>0.8) are very close to the actual locations of ligand atoms, with an average distance of 0.62 Å. This finding indicates that our model’s predicted high binding probability regions closely correspond to the actual locations of the ligand heavy atoms. Thus, our model shows great promise for predicting ligand poses by aligning them with the predicted probability density envelope.
We also applied FeatureDock to predict binding probability density envelopes on different conformational states of the same protein. As shown in Supplementary Fig. 5, the probability density envelopes of the inactive and active CDK2 exhibit substantial differences in the region proximal to Tyr15, located at the glycine-rich loop (G-loop, 11–16). Consequently, an allosteric ligand binding near Tyr15 may hinder the structural transition from the inactive to the active state, thereby inhibiting the CDK2 activation. This prediction is consistent with the reported ANS allosteric binding site44 located between the ATP binding pocket and C-helix which can inhibit cyclin binding.
Lastly, we further demonstrated the robustness of FeatureDock’s probability density envelop by comparing it with Vina, AutoDock4 and DiffDock on the benchmark PDBBind v2020 refined dataset. Specifically, among the 1326 protein structure clusters (Supplementary Figure 1), we randomly selected 10% of the protein structures, resulting in a test dataset containing ~660 protein-ligand complex structures. For ligand compounds in the test dataset, we first generated predicted probability density envelopes using FeatureDock. For comparison, we used 3 other docking methods, DiffDock, Vina, and AutoDock4, to predict the top 10 binding poses for each compound. We then estimated the binding probabilities at grid points in the binding pocket based on the occupancy of these binding poses. The resulting probability density envelopes enabled a more comprehensive performance evaluation of FeatureDock compared to the other three docking methods. We first used the metric of binary cross-entropy between the predicted probability density envelopes and the ligand occupancy in each protein-ligand structure (referred to as the “ground truth”). As shown in Supplementary Fig. 17A, FeatureDock achieves the lowest cross-entropy loss relative to the ground truth binding pose, outperforming all other methods and demonstrating its robustness in pose prediction. In addition, we compared these methods using the F1 Score, a metric that evaluates the quality of predicted docking results by balancing both precision and recall. This measure captures how effectively each method identifies binding poses that closely match the ground truth while minimizing false positives. Specifically, we assigned “binding” and “non-binding” labels based on probability density envelopes at different cutoffs to compute the F1 Score. As shown in Supplementary Fig. 17B, DiffDock demonstrates the best overall performance, while both FeatureDock and DiffDock outperform Vina and AutoDock4. Notably, when the cutoff reaches 0.9, FeatureDock and DiffDock achieve comparable performance.
Identifications of chemical features contributing most to ligand binding
Our FeatureDock tool generates attention maps for each input chemical feature using the attention weights from the Transformer architecture (see Supplementary Fig. 3 and Eq. (1)). Each attention map records the contributions of a specific chemical feature in predicting the binding probabilities of all grid points in the binding pocket. For example, Fig. 2B visualizes the attention maps of two representative chemical features (hydrophobicity and polar residues) of grid points in the binding pocket for an inactive form of CDK2 structure (PDB ID: 1B3845). In Fig. 2A, we report the average attention weights per given amino acid surrounding the binding pocket. Among all the hydrophobic residues, Phe82 and Phe80 contribute the most to the ligand binding, with contributions of 2.6% and 2.0%, respectively (top panel of Fig. 2A). The attention weights from our FeatureDock tool is consistent with the previous results obtained from the pharmacophore model46. In this pharmacophore model, hydrophobic pharmacophores containing Phe82 and Phe80 emerged as frequently occurring features based on multicomplex-based analysis constructed on 124 CDK2 cocrystal structures. For polar amino acids surrounding the binding pocket, Gln85 and Gln131-Asn132 are predicted by our FeatureDock to mostly contribute to the ligand binding (bottom panel of Fig. 2A). Among these residues, Gln85 has also been predicted by the previous pharmacophore modeling study46 as a favorable interacting residue for pharmacophore binding. Moreover, previous Molecular Dynamics (MD) simulation studies47,48 also pinpointed Gln85 as a key residue in designing ligands selectively binding to CDK2. Specifically, replacing the chemical groups on the ligand near Gln85-Lys89 with more electronegative ones enhances ligand selectivity for CDK2 over CDK7. This previous finding supports our observation that Gln85 contributes as large as 3.9% to the ligand binding. In addition, while Gln131 and Asn132 currently show low statistical occupancy as hydrogen bond acceptors or donors in the pharmacophore model46, we anticipate that designing ligands to interact with these two residues via favorable hydrogen bonding interactions could improve the CDK2 ligand selectivity, as both of these residues show high contributions in our attention analysis (bottom panel of Fig. 2A).

A Heatmap of property contribution to the amino acids of interest by averaging attention weights of grid points around each amino acid. The picked residues frequently form contact with ligands in cocrystal structures. The text of hydrophobic residues and polar residues are colored in magenta and green, respectively. The range of attention weights is [0, 1]. B The attention weight map of different properties of the spatial grid points. Regions in darker color have higher contribution values in the attention analysis.
FeatureDock outperforms DiffDock, Smina and AutoDock Vina in differentiating strong and weak inhibitors
As discussed in the Introduction section, distinguishing between strong and weak inhibitors remains challenging for docking algorithms during virtual screening, as all inhibitors tend to fit reasonably well in the binding pockets. In this section, we demonstrate that our FeatureDock tool outperforms traditional docking methods (Smina22 and AutoDock Vina21), as well as the recently developed diffusion-based generative model (DiffDock28) in this task for an inactivated form of CDK2 and the ACE receptor. In FeatureDock, we defined a scoring function based on Eq. (2), which assesses the agreement between the locations of ligand atoms and the predicted probability density envelopes. To optimize the ligand’s position and orientation in the binding pocket, we implemented an optimization procedure using the L-BFGS-B algorithm to minimize the objective function defined in Eq. (3). For additional details on our optimization protocol and the preparation of query structures and compound libraries for virtual screening, please refer to Methods Sections “Scoring functions and virtual screening” and “Preparation of query structures and compound libraries”. To evaluate the performance of virtual screening, we used two metrics: KL divergence and AUC. A higher KL divergence indicates that the model can more effectively distinguish strong and weak inhibitors. Similarly, a higher AUC demonstrates the scoring function’s effectiveness in ranking strong inhibitors above weak ones. The parameters used in benchmark docking methods are detailed in Methods Section “Parameters used in the benchmark docking programs”.
Inactivated form of CDK2
We compiled a library containing 147 compounds followed by the filtering rules described in the Methods Section “Preparation of query structures and compound libraries”. As shown in Fig. 3A, FeatureDock achieved the largest KL divergence (0.67) in differentiating strong inhibitors from weak inhibitors, outperforming DiffDock (0.39), Smina (0.04), and Vina (0.04). The peaks of score distributions of FeatureDock showed better separation compared to DiffDock in Fig. 3A. These results suggest that we can use the FeatureDock scoring function to select reliable candidates which achieve higher scores in the virtual screening. Moreover, the peak of strong inhibitors is mixed with that of weak inhibitors in both Smina and AutoDock Vina, which explains the high false positive ratios caused by the scoring functions used in these two docking methods. Furthermore, as shown in Fig. 3B, the AUC showed that FeatureDock (0.74) and DiffDock (0.76) can rank strong inhibitors higher than the weak ones, while the binding affinity scores used by Smina (0.43) and Vina (0.43) ranked strong and weak inhibitors in this library worse than random guess. These two-evaluation metrics both support the effectiveness of FeatureDock’s scoring function (defined in Eq. (2)) during virtual screening. Furthermore, since outlier false positive may affect the KL divergence, e.g., an incorrect prediction can also increase the KL divergence, we conducted additional experiments on a filtered library (113 compounds) by removing these outliers. As shown in Supplementary Fig. 18, even after this refinement, FeatureDock’s KL divergence and AUC values remain higher than those of the other benchmark methods.

A KL divergence of score distribution on strong inhibitors and weak inhibitors from a filtered CDK2 bioassay dataset which contains 147 compounds. B AUC of scoring functions on the 147-compound library. C True positive (the blue boxed) and true negative (the orange boxed) examples of predicted poses overlaid with the predicted probability density envelope. The colorbar represents colors of different probability values. Compound atoms in the regions of p ≥ 0.95, 0.90 ≤ p < 0.95 and 0.80 ≤ p < 0.90 are colored in green, yellow, and red respectively. Surrounding protein structures are shown in white ribbon, with important binding residues Phe80 and Phe82 highlighted in white sticks.
In FeatureDock, we simultaneously score compounds and optimize their binding poses using the L-BFGS-B optimization algorithm. In Fig. 3C, we show the optimal ligand poses obtained from FeatureDock overlaid with their predicted probability density envelopes for one strong inhibitor (CHEMBL402158) and two weak inhibitors (CHEMBL3421971 and CHEMBL3798066) of CDK2. In the FeatureDock pose of the strong inhibitor (CHEMBL402158), its 1H-indazole group occupies the highest probability regions (p ≥ 0.95) within the hydrophobic pocket near Phe80 and Phe82 of CDK2. The remaining portion of this strong inhibitor extends into adjacent regions with similarly high probability values (0.90 ≤ p < 0.95). As a result, CHEMBL402158 exhibits a high score in FeatureDock, aligning with its experimentally measured strong IC50 of 7 nM (Left panel of Fig. 3C). For the weak inhibitor CHEMBL3421971 (Middle panel of Fig. 3C), its benzene and piperazine groups also occupy the highest probability region (p ≥ 0.95) near the hydrophobic residues Phe80 and Phe82. However, the linker atom connecting these function groups to the rest of the compound bypasses a region with substantially lower probabilities (as indicated by the red-labeled atom in the Middle panel of Fig. 3C). As a result, FeatureDock assigns a lower score to CHEMBL3421971, consistent with its experimentally measured IC50 of approximately 13.7 µM. As for the other weak inhibitor, CHEMBL3798066, a significant portion also occupies a relatively low probability region (atoms labeled in red in the Right panel of Fig. 3C), resulting in a weak inhibitor (IC50 = 11 µM). These examples indicate that FeatureDock provides not only an accurate scoring function, but also meaningful optimal ligand poses, making it promising for virtual screening.
To further assess the accuracy of pose prediction from FeatureDock, we validated its predicted poses by comparing to the co-crystal structures for 11 native ligands of CDK2 (the inactivated form) that have cocrystal structures in PDBBind. The posing performance is evaluated by root mean square distance (RMSD) between the native pose and the predicted pose. An RMSD below 2 Å is usually considered to be a good prediction in docking methods. After pose optimization described in Methods Section “Scoring functions and virtual screening”, the RMSD converged after picking 4 top-scored poses for each ligand in FeatureDock. The average and median RMSD achieved by our method are 2.4 Å and 2.1 Å, which is significantly better than randomly choosing poses centered to the predicted regions while not optimized via soft alignment based on the probability density envelope. Examples of predicted poses in Fig. 4 right panel show the alignment performance.

The query space of each protein refined by protein residues within [1, 6] Å away from the heavy atoms of its native ligand. The native rotamer was used in the posing process. Left panel: Distribution of root mean square distances (RMSDs) between predicted poses and native poses. Right panel: Examples of predicted poses. The ground truths and the predictions are presented in white sticks and magenta sticks, respectively.
ACE receptor
We followed the filtering rules described in Methods Section “Preparation of query structures and compound libraries” and compiled a compound library containing 94 compounds. On ACE, FeatureDock also achieves the highest KL divergence (0.30) (Fig. 5A) and AUC (0.69) (Fig. 5B), outperforming DiffDock, Smina and AutoDock Vina. A relatively higher probability cutoff of 0.90 was used to re-score the compound occupancies for ACE based on the result of hyperparameter scanning (Supplementary Fig. 9). For the details of how to select the proper probability cutoff as a scoring hyperparameter for different protein systems, please refer to the Methods Section “Scoring functions and virtual screening”. We visualized one true positive prediction and two true negative prediction to analyze the performance of scoring and pose prediction. As depicted in Fig. 5C, the true positive prediction CHEMBL100413 occupied the regions of p ≥ 0.95 close to Tyr520, which is one of the lisinopril-binding residues49,50. Furthermore, the predicted pose of CHEMBL100413 exhibits the correct orientation to form the hydrogen bond with Tyr520, which explains its high potent to inhibit ACE. The true negative prediction CHEMBL5220968 occupied the same region of p ≥ 0.95 except that the compound sticks out of the probability envelope due to the compound geometric constraint, leading to a lower score. Another true negative prediction CHEMBL5177969 achieved an even lower score because it totally missed the regions of highest probabilities.

A KL divergence of score distribution on strong inhibitors and weak inhibitors from a filtered ACE bioassay dataset which contains 94 compounds. B AUC of scoring functions on the 94-compound library. C True positive (the blue boxed) and true negative (the yellow boxed) examples of predicted poses overlaid with the predicted probability density envelope. Compound atoms in the regions of p ≥ 0.95, 0.90 ≤ p < 0.95 and p ≤ 0.80 are colored in green, yellow, and red respectively. The colorbar represents colors of different probability values. Surrounding protein structures are shown in white ribbon. The active site (NEXXH motif) and the Tyr520 residue are shown as white sticks.
Discussion
FeatureDock is a structural-based molecular docking method that both predicts the binding poses and evaluates the binding potency of compounds based on their alignment with the probability density envelopes. Since FeatureDock also exhibits scoring power, to demonstrate its potential advantages and limitations, we performed additional experiments to directly compare scoring power of FeatureDock with other machine learning-based scoring functions. Specifically, we conducted the comparisons between FeatureDock and RF-score51 on entries from the PDBBind v2020 refined dataset containing druglike molecules of molecular weight between 400 and 600. As shown in Supplementary Fig. 19A and 19B, RF-Score outperforms FeatureDock, achieving a higher Pearson correlation of 0.647 than FeatureDock (0.408), in predicting the experimental binding affinity. This result is expected, as RF-Score is trained on these binding affinities, whereas FeatureDock is a molecular docking method, and its training dataset does not contain any experimental binding affinity data. Even though RF-Score performs well on predicting binding affinities of the complexes from PDBBind v2020, we anticipate that its performance will be greatly limited when applied to predict binding affinities of chemical compounds outside its training dataset. To test this hypothesis, we designed a control experiment: we applied RF-Score-VS52, a virtual screening variant of RF-Score but trained on the DUD-E53 dataset rather than PDBBind v2020 dataset, to repeat the compassion. As shown in Supplementary Fig. 19C, RF-Score-VS demonstrates poor performance in virtual screening of the PDBBind dataset, raising concerns about the generalization capability of such machine-learning-based scoring functions.
In FeatureDock, we utilize the L-BFGS-B optimization algorithm to find the ligand pose that best fits to the predicted probability density envelope, and we acknowledge that this optimization process could be trapped into local energy minima. To address this issue, we perform independent optimization runs initiated from different compound rotamers and using different random seeds. This strategy has successfully identified the correct binding pose for CDK2 (see Fig. 4). In the future, to further improve the efficiency of pose sampling and optimization, one could incorporate E3-equivariant diffusion models to generate binding positions based on the probability density envelopes.
When applying FeatureDock to virtual screening, it is important to select a proper probability cutoff in the scoring function. We recommend choosing its value by scanning the probability cutoff as a hyperparameter on the compound bioassay data and pick the best cutoff that achieves the highest AUC. Intuitively, for more rigid pockets that do not allow multiple binding modes, a higher probability cutoff can help refine the search space, focusing only on regions containing grid points with confident ligand binding predictions. For example, a higher cutoff value was chosen for ACE than CDK2 in the Results Section “FeatureDock outperforms DiffDock, Smina and AutoDock Vina in differentiating strong and weak inhibitors”, which is consistent with the observation that the ACE ligand binding pocket has lower flexibility compared to that of CDK2, as evaluated by higher B-factor values for ACE (Supplementary Fig. 11). When the bioassay data is not available, we would suggest using a balanced probability cutoff (e.g. ({cutoff}=0.90)) to guarantee that high probability regions are prioritized to be occupied, while remaining a probability density envelope that can represent and cover the binding pocket adequately. This cutoff has shown effective in the virtual screening of both CDK2 and ACE (Supplementary Figs. 8 and 9).
One challenge facing deep learning-based docking methods is their tendency to generate “invalid” ligand poses54. This occurs because these methods often produce poses with unphysical chemical bonds or steric clashes between the protein and ligand, as they do not explicitly include physical interactions during training and pose prediction. To address this challenge, we introduced a post-analysis step to identify and remove unphysical poses generated by FeatureDock and DiffDock (Supplementary Fig. 10). In this step, we utilized AutoDock Vina for local optimization to identify steric clashes and other unphysical interactions that result in unfavorable affinity scores. Specifically, we rejected poses that with affinity scores larger than 0. It is important to note that we used docking affinity only to identify steric clashes, not to re-pose the conformations generated by FeatureDock or DiffDock. In the future, one could directly introduce a physical interaction term to avoid steric clashes in the objective function of the pose optimization process in the FeatureDock.
FeatureDock employs a two-step virtual screening pipeline. The probability prediction and scoring per structure is fast, while the pose prediction is relatively slow because we haven’t utilized efficient searching algorithms in this step. As shown in Supplementary Fig. 20, we conducted a computational efficiency test by randomly selecting 100 structures from the PDBBind v2020 refined set and reported the average processing time per structure. To achieve a fair comparison, the speed tests were all conducted on Intel(R) Core (TM) i7-10510U.
In Conclusion, we introduced FeatureDock, a feature-based Transformer framework for predicting and scoring protein-ligand binding poses. FeatureDock extracts the physicochemical features of the protein’s local environment and utilizes a transformer encoder to predict probability density envelopes for given protein pockets. These predicted probability density envelopes help determine the binding preference of the ligand at grid points within the protein pockets. Compared to FNN and ResNet architectures, Transformer encoders demonstrate superior performance, achieving higher and more stable prediction accuracy throughout comprehensive model capacity tests. In addition, the attention mechanism in the Transformer encoder further shows potential for assisting rational drug design by elucidating the contribution of each chemical feature to the final predicted ligand binding probabilities. Using the predicted probability density envelopes, we designed a custom scoring function and a ligand-position optimization algorithm to score and predict the protein-ligand binding poses. In virtual screening of drug-like compounds, FeatureDock outperforms DiffDock, Smina and AutoDock-Vina in distinguishing strong inhibitors from weak ones for both the inactivated form of CDK2 and ACE systems. FeatureDock also effectively predicts their binding poses accurately. We expect that FeatureDock holds potential to be widely applied in virtual screening, aiding in drug design.
Methods
We outline the pipeline for our FeatureDock framework in Fig. 6. Initially, the ligand-binding pockets of protein complexes in the PDBBind v2020 refined set55 are discretized into grid points, embedded using 3D-invariant FEATURE representations (Fig. 6A, B). We then train a Transformer encoder model as shown in Fig. 6C. After training, our model predicts the binding probabilities of grid points in the query pockets of a given apo protein structure, forming probability density envelopes (Fig. 6D). Using these probability density envelopes, we designed a scoring function combined with a position optimization algorithm to predict and score the binding poses (Fig. 6E). We will describe these steps in detail below:

A Collect protein-ligand complexes from PDBBind v2020 refined set. B Extract and featurize protein local environments around grid points in the ligand-binding pocket and then label the grid points as either binding or non-binding with the ligand. C Train the Transformer Encoder to predict the ligand-binding probability of each grid point. D Predict the probability density envelope for the query space in apo proteins using the trained model. Grid points with darker color are more likely to be occupied by compounds. E Apply the predicted probability density envelope to virtual screening and pose prediction.
Dataset curation
Protein representation and data labeling
The training dataset was curated from the PDBBind v2020 refined set55 (Fig. 6A), comprising 5,316 high-quality protein-ligand complex structures that have been cleaned, fixed, and properly protonated from raw structures deposited in RCSB PDB56. Notably, ligands in the refined set are all non-covalently binding to the proteins. We extracted the convex hull formed by protein residues within 6 Å around each ligand. This space was discretized to grid points spaced by 1 Å. The grid points too close to (<1 Å) or too far away from (>6 Å) the protein atoms were removed to reduce the dataset size because ligand atoms are less likely to show up in these regions.
Protein-ligand conformations are unstructured data compared to image tensors, thus requiring delicate data preprocessing before being fed into neural networks. Having a good protein representation is one of the determining factors to make sure that the model learns meaningful information. In our previous work focused on predicting protein-RNA interactions42, FEATURE vectors41 have been proven descriptive enough to capture protein local environments. Within each spheric shell, FEATURE vectors encode 80 physicochemical properties (Supplementary Table 1) across various hierarchies, including atomic level, residue-level, and secondary structural level information. Since FEATURE vectors are invariant to rotational and permutational changes of protein structures, there is no requirement of dataset augmentation to make models robust against these operations. Furthermore, we will show that fine-tuning of the model on a specific protein family is not necessary because the model trained on various proteins has the generalization ability to predict novel structures, as shown in the Results Section “Model training and selection”.
By default, the local environment surrounding each grid point was characterized by 6 concentric shells (Fig. 6B). Centered at each grid point, these shells were defined with a width of 1.25 Å each, capturing properties in a local environment up to a radius of 7.5 Å. FEATURE vectors, were then applied to these concentric shells with the 80 pre-defined physicochemical properties. This featurization resulted in a 6×80 tensor for each grid point. Grid points were assigned with labels related to the protein-ligand binding properties. Here we chose a categorical label, which tells whether there is a ligand heavy atom within 1.5 Å of the grid point (Fig. 6B).
After extracting features from existing protein-ligand cocrystal structures and curating a labeled dataset for supervised learning, the Transformer encoder can be trained (Fig. 6C) and then applied to predict which space is more likely occupied by ligands in a data-driven manner for novel apo structures (Fig. 6D). Later, the predicted probability density envelope formed by the grid points and probabilities will be used in the scoring function during virtual screening (Fig. 6E). Notably, as the probability density envelope is derived from the grid point space, we conducted a sensitivity analysis to assess its dependence on grid points selection. The results indicate that the probability density envelopes are not significantly influenced by the choice of grid points. For further details, please refer to Supplementary Note 1 and Supplementary Fig. 12.
Dataset split
To avoid trivial queries and predictions during the model validation, we split the whole curated dataset (containing 4515 structures after pre-processing) into training/validation/test set based on their sequence similarities. Structures were clustered to 1326 structure clans (Supplementary Fig. 1) based on 90% MMseq257 protein sequence similarity fetched from RCSB PDB56. 10% of the protein clusters were randomly selected as the validation set and the remaining were used for training. This split method guarantees that homogeneous structures exist in either the training or validation dataset but not both. Furthermore, this split strategy mitigates potential data leakage caused by similar ligands showing up in both the training and validation sets (see Supplementary Note 2 and Supplementary Fig. 13 for details), compared to a random split. By adopting this dataset split strategy, the evaluation metrics of the validation set can better reflect the model performance and avoid overfitting. During the validation of model generality (the leave-out models), models were trained on the dataset after leaving out one certain type of protein (e.g., CDK2) and its homogeneous structures (sequence identity ≥ 90%) to mimic the scenario of applying the trained model to predict novel protein structures. After validating that our model could achieve good performance on predicting novel structures, we trained full models with all structure clusters and used the full-model predictions in the model application of virtual screening and pose prediction.
Data resampling for mitigating class imbalance
In this section, we discuss the class imbalance in available training protein complex structures. One source of imbalance comes from the fact that some protein families are better studied than others, leading to the structure abundance difference. For example, ACE proteases have <10 structures in the PDBBind refined dataset, CDK2 and CDK2/Cyclin have ~20 structures, while many other proteins have only one single structure (Supplementary Fig. 1). Another source comes from the fact that grid points with negative labels (non-binding regions) are more abundant than those with positive labels (binding regions) because we selected a relatively large pocket compared to the regions that the ligand occupy in the step of dataset curation. We adopted two data resampling strategies to address the class imbalance issue. The first is to sample each protein cluster uniformly instead of sample each protein structure uniformly. The model can therefore have better generalization capability by sampling local environments from rare protein clusters more frequently. The second is to balance the abundance of different labels by under-sampling negative datapoints and over-sampling positive datapoints.
Neural network architectures
FeatureDock used the Transformer encoder (Fig. 6C) to predict the ligand binding probabilities of grid points because it consistently outperformed Feed Forward Neural Network and ResNet across various model sizes. To elucidate that Transformer encoders could achieve better performance, we compared different neural network architectures at different model sizes, and evaluated their performances by the loss, area under ROC (AUC), F1 Score and Matthews correlation coefficient (MCC) on the validation set.
Transformer encoder
Transformer43 was first proposed in 2018 for natural language processing (NLP) tasks. It pioneered the use of attention mechanism to capture the temporal correlations in token sequences, enabling the efficient parallel computing compared to traditional recurrent neural networks, long short-term memory (LSTM)58, etc. The success of Transformer has also inspired significant advancements in the field of computer vision (CV). Various transformer-based models, e.g., vision transformer (ViT)59 and swin transformer60, have been developed, outperforming most of traditional CNNs in image classification tasks. Compared to convolutional operations that capture local spatial correlation, the attention mechanism in Transformer calculates all-to-all correlations among the input features. Transformer has also gained its popularity in the drug discovery field. For example, Transformer has been adopted to encode protein sequences and generate SMILES of potential drugs, or to study drug-target interactions (DTI)61.
We leveraged the Transformer encoder to predict the ligand-binding probability of each grid point based on the physicochemical properties of its local environment. Specifically, we conceptualized each physicochemical property as analogous to a word (Fig. 7). Values of the same property in different concentric spheritic shells (total of 6 shells) were aggregated into a single word (consisting of 6 characters in each word). The class token59,62 was concatenated with the 80 properties to form an 81-word sentence ({{boldsymbol{h}}}_{{boldsymbol{0}}}) for the classification purpose. The 81-word sentence was additionally encoded with positional embeddings and fed to the encoder blocks. The final embedding of the class token can be viewed as the representation of the grid point after propagating information of input properties. It was then fed into the Softmax layer which outputs the binding probability.

80 physicochemical features of each grid point and the class token are fed into the Transformer encoder. Each feature is a 6-dimensional vector that encodes certain properties of the grid point’s local environment, and the class token is used to predict the probability of this grid point being occupied by ligand atoms.
Here we denote the input of each encoder block (Supplementary Fig. 3 and Eq. (1)) as ({{boldsymbol{h}}}_{{boldsymbol{i}}}{boldsymbol{in }}{{mathbb{R}}}^{{boldsymbol{l}}{boldsymbol{times }}{boldsymbol{d}}}) and the output as ({{boldsymbol{h}}}_{{boldsymbol{i}}{boldsymbol{+}}{boldsymbol{1}}}{boldsymbol{in }}{{mathbb{R}}}^{{boldsymbol{l}}{boldsymbol{times }}{boldsymbol{d}}}), which will then serve as the input of the next block. (l) is the sequence length ((l=81)), and (d) is the dimension of each word (i.e., chemical property) in the sequence. In each block, the input ({{boldsymbol{h}}}_{{boldsymbol{i}}}) is projected to the query (({boldsymbol{Q}})), key (({boldsymbol{K}})), value (({boldsymbol{V}})) matrices, parameterized by ({{boldsymbol{W}}}_{{boldsymbol{Q}}},{{boldsymbol{W}}}_{{boldsymbol{K}}},{{boldsymbol{W}}}_{{boldsymbol{V}}}{boldsymbol{in }}{{mathbb{R}}}^{{boldsymbol{d}}{boldsymbol{times }}{{boldsymbol{d}}}^{{boldsymbol{{prime} }}}}). The attention mechanism utilizes ({boldsymbol{Q}}), ({boldsymbol{K}}) ({boldsymbol{in }}{{mathbb{R}}}^{{boldsymbol{l}}{boldsymbol{times }}{{boldsymbol{d}}}^{{boldsymbol{{prime} }}}}) to calculate a word-to-word weight matrix ({boldsymbol{S}}{boldsymbol{in }}{{mathbb{R}}}^{{boldsymbol{l}}{boldsymbol{times }}{boldsymbol{l}}}), where ({boldsymbol{S}}(j,{k})) represents the weight of word (k) contributing to word (j). The updated hidden state ({{boldsymbol{h}}}_{{boldsymbol{i}}{boldsymbol{+}}{boldsymbol{1}}}^{{boldsymbol{{prime} }}}{boldsymbol{in }}{{mathbb{R}}}^{{boldsymbol{l}}{boldsymbol{times }}{{boldsymbol{d}}}^{{boldsymbol{{prime} }}}}) (({d}^{{prime} }) is the dimension of the hidden state) is a weighted combination of the value matrix ({boldsymbol{V}}), given the attention weight matrix ({boldsymbol{S}}). The final output of the block ({{boldsymbol{h}}}_{{boldsymbol{i}}{boldsymbol{+}}{boldsymbol{1}}}) will be calculated using ({{boldsymbol{h}}}_{{boldsymbol{i}}{boldsymbol{+}}{boldsymbol{1}}}^{{boldsymbol{{prime} }}}) and ({{boldsymbol{h}}}_{{boldsymbol{i}}}) via residual connection. The attention weight from physicochemical properties to the class token can be used to further enhance model interpretability by visualizing contributing weights (see Fig. 2 for examples). In implementation, we utilized multi-head attention that projects the queries, keys and values (h) times parallelly, and concatenated their outputs (see Supplementary Fig. 3 for details). When calculating the contribution of features to ligand binding, we extracted and averaged the weight matrices of attention heads in the final encoder block.
Benchmark Model 1: Feed Forward Neural Network (FNN)
Feed forward neural network (FNN) is a stack of fully connected layers with different number of neurons. Each layer performs a linear combination of input features followed by a nonlinear activation function performed on each hidden feature. At the end of the neural network, a Softmax layer is applied to provide the probabilities of assigning each input data to different categories. In our scenario (Supplementary Fig. 2A), the 6 × 80 input features corresponding to each grid point are serialized as a 480-dimensional vector ({{boldsymbol{h}}}_{{boldsymbol{0}}}), which is then passed into FNN.
Benchmark Model 2: ResNet
CNNs use convolutional operations to capture localized patterns in images and have exhibited robustness in image classification tasks. For example, pioneering CNN architectures like AlexNet63 achieved a prominent top-1 accuracy of 63.3% and showed a much lower top-5 error rate by at least 10% compared to the other models in 2012 ImageNet contest. Subsequently, in 2014, ResNet64 was introduced, revolutionizing CNNs by introducing residual connections. The residual connection enables the identity mapping across different convolutional blocks, significantly enhancing the robustness of larger deep learning models. Furthermore, residual connections alleviated the gradient vanishing/explosion issue that otherwise may occur in standard neural network models.
As shown in Supplementary Fig. 2B, the 6 × 80 features were reshaped to a 1 × 6 × 80 tensor ({{boldsymbol{h}}}_{{boldsymbol{0}}}) and fed into ResNet blocks. In each ResNet block, the input state ({{boldsymbol{h}}}_{{boldsymbol{i}}}{boldsymbol{in }}{{mathbb{R}}}^{1{rm{x}}6{rm{x}}80}) was filtered by the 1 × 2 × 80 convolutional kernels and converted to the hidden state ({{boldsymbol{h}}}_{{boldsymbol{i}}{boldsymbol{+}}{boldsymbol{1}}}) with the same dimension. This kernel shape can capture spatial correlation among properties in two adjacent concentric shells and allows the information from outer shells to pass to inner shells when more residual blocks are added on.
Hyperparameters and fine-tuning
We used cross-entropy loss as the objective function of our models. The training hyperparameters are summarized in Supplementary Table 2. The learning rate was set as 0.01, with a decay factor of 0.5 when the validation loss reached a plateau, and the minimum threshold was 10−6. The optimizing method was AdamW optimizer65 with betas (0.9, 0.999). To better reflect the true model capacity, no weight decay was applied during model capacity testing and architecture selection procedures. The best model was recorded based on the validation loss to avoid overfitting. We trained each model by 5 random above-mentioned training/validation splits to get validation statistics and select the best model architecture with the most suitable capacity.
The default training epochs was set to 30 in the leave-out model training and 50 in the full model training. The default batch size was 5000, which meant sampling 5 structures from each structure cluster and sampling 1000 class-balanced datapoints from each structure. Benefitted from this dynamic resampling process, the model tended to iterate over different structures of each structure cluster as the training went on. The models were trained on NVIDIA A100 SXM4 80GB.
Scoring functions and virtual screening
We encoded the grid-point coordinates ({boldsymbol{p}}) together with ligand coordinates ({boldsymbol{q}}) in our scoring function (Eq. (2)). ({boldsymbol{p}}) has the shape of ((N,3)), where (N) represents the number of grid points. ({boldsymbol{P}}left({boldsymbol{p}}right)) represents the predicted probabilities of the grid points. ({boldsymbol{q}}) has the shape of ((M,3)), where (M) means the number of heavy atoms in the given compound. ({boldsymbol{T}}) is the transformation matrix that incorporates translational and rotational operations in the three-dimensional Euclidean space. The scoring function rewards compound poses that can occupy space of higher predicted probability. In the following equations, ({{boldsymbol{p}}}_{{boldsymbol{i}}}) represents the coordinate of the ith grid point, and ({{boldsymbol{q}}}_{{boldsymbol{j}}}) represents the coordinate of jth ligand heavy atom.
The binding position of any compound can be optimized by L-BFGS-B to minimize the objective function (Eq. (3)). It is worth noting that the objective function considers distance-weighted probabilities of all grid points given a certain probability cutoff, while the scoring function rescales the objective function by only considering the neighboring (≤1.5 Å) grid points ({{boldsymbol{N}}}_{{boldsymbol{p}}}left({{boldsymbol{Tq}}}_{{boldsymbol{j}}}right)). We used ({probability_cutoff}=0.50) in the objective function to allow a larger compound explorable region and used a different probability cutoff to re-score the optimized poses for different protein systems. During scoring, we suggest using the best probability cutoff evaluated on AUC when the bioassay results are available for the query protein. Please refer to Results Section “FeatureDock outperforms DiffDock, Smina and AutoDock Vina in differentiating strong and weak inhibitors” for the details of the above-mentioned optimization and scoring method in selecting strong inhibitors from compound libraries for inactive CDK2 and ACE. When the bioassay results are unavailable, it is suggested to choose the probability cutoff based on the pocket properties, which has been elaborated in the Discussion section.
To predict binding poses using the probability density envelope, we first generated up to 20 conformers for each compound using RDKit (with pairwise RMSDs between these conformations are larger than 1 Å after molecular alignment). useExpTorsionAnglePrefs and useBasicKnowledge were enabled during sampling, and the generated conformers were further optimized by the MMFF66 force field. Each ligand conformer was initialized by 500 random rotations. An utmost of 10,000 optimization results per compound were finally clustered by DBSCAN67,68 method based on their RMSDs to reduce the number of aligned poses. The proposed clusters were sorted by the average score of cluster members. The purposes of massive initial samplings followed by a clustering process are to avoid potential local minima during L-BFGS-B optimization, and to reduce the number of proposed binding poses. We combined sampling and clustering of three different random seeds to make the top-proposed clusters more robust.
Preparation of query structures and compound libraries
Compound libraries used in virtual screening were fetched from the CHEMBL8 database. The compounds with IC50 less than 10 nM are considered as strong inhibitors and those with IC50 values greater than 10 µM are considered as weak inhibitors. The filtered compound library from CHEMBL demonstrated a low compound similarity to ligands in PDBBind refined dataset (see Supplementary Note 3 and Supplementary Fig. 14 for details). To mimic a blind virtual screening, we further removed compounds that share >95% fingerprint Tanimoto similarity with ligands in the PDBBind cocrystal structures to curate compound libraries different from native ligands used in training. This setting helps validate the model’s ability to select novel compounds that are different from existing ligands from a pre-selected drug-like compound library.
An inactive form of CDK2
The cleaned apo structure of 1B38 was used as the reference protein structure of CDK2 during virtual screening. The query space (Supplementary Fig. 6A) used in our method is formed by grid points within 5 Å away from native ligands of 11 pre-aligned CDK2 structures.
We filtered compounds with molecular weight greater than 400 Da from CHEMBL301 which consists of CDK2 bioassay results with IC50 values. Compounds in CHEMBL301 with bioassay descriptions explicitly reporting to be tested on CDK2/Cyclins are removed because they bind to a different protein conformation caused by cyclin activation. The above filtering rules result in a 147-compound virtual screening library for the CDK2. The molecular property distributions of strong inhibitors and weak inhibitors in this library are similar (Supplementary Fig. 6B).
ACE
The cleaned apo structure of 3BKL was used as the reference protein structure of ACE. The query space (Supplementary Fig. 7A) used is formed by grid points within 5 Å away from native ligands of 4 pre-aligned ACE structures that share a similar scaffold (2OC2, 3BKK, 3BKL and 6F9U), but it is large enough to cover all 7 native ACE ligands in the cocrystal structure dataset.
We filtered compounds with molecular weight greater than 400 Da and less than 600 Da from CHEMBL1808 which consists of ACE bioassay results with IC50 values. We further used the following criteria to filter out compounds less likely to be drugs and make the compound properties match in the meanwhile (Supplementary Fig. 7B): logP ≤ 5, number of hydrogen donors ≤5, number of hydrogen acceptors ≤10, number of rotatable bonds ≤5. The above filtering rules result in a 94-compound virtual screening compound library.
Parameters used in the benchmark docking programs
Protein structures were prepared using AutoDockTools1.5.7 for Smina and AutoDock Vina. The query box of inactive CDK2 has the size of 18 Å × 16 Å × 16 Å, centered at [1.38, 26.15, 9.40]69 (Supplementary Fig. 4). The query box of ACE has the size of 18 Å × 22 Å × 24 Å, centered at [43, 45, 44]. These two docking boxes are large enough to cover all ligands from cocrystal structures in the curated dataset. sdf files of compounds are generated from SMILES using RDKit. pdbqt files used in docking softwares were converted from sdf files using openbabel70. The parameters used in Smina and in AutoDock Vina are exhaustiveness = 8, num_modes = 20.
The default parameters of DiffDock were used to generate compound conformations and positions during the virtual screening: inference_steps = 20, samples_per_complex = 40, batch_size = 10 actual_steps = 18, no_final_step_noise = True.


Responses