Development and evaluation of deep learning models for cardiotocography interpretation

Introduction
Intrapartum cardiotocography (CTG) is a screening technique that is widely used to monitor fetal well-being by recording the fetal heart rate (FHR) along with the maternal uterine contractions (UC) during labor. Although CTG is routinely used in medical practice, the use of continuous intrapartum fetal monitoring is associated with a high false-positive rate. This has led to an increase in Cesarean section and operative vaginal delivery rates with limited improvements in neonatal outcomes1 due to the subjectivity of current methods2,3 and intra-observer variability4. While maternal and neonatal mortality is influenced by multiple factors beyond the scope of CTG interpretation alone, the limitations of current CTG interpretation methods are exacerbated in facilities where access to skilled interpreters is limited5,6. Figure 1 highlights the aforementioned challenges with visual CTG interpretation and the relevant clinical use case.
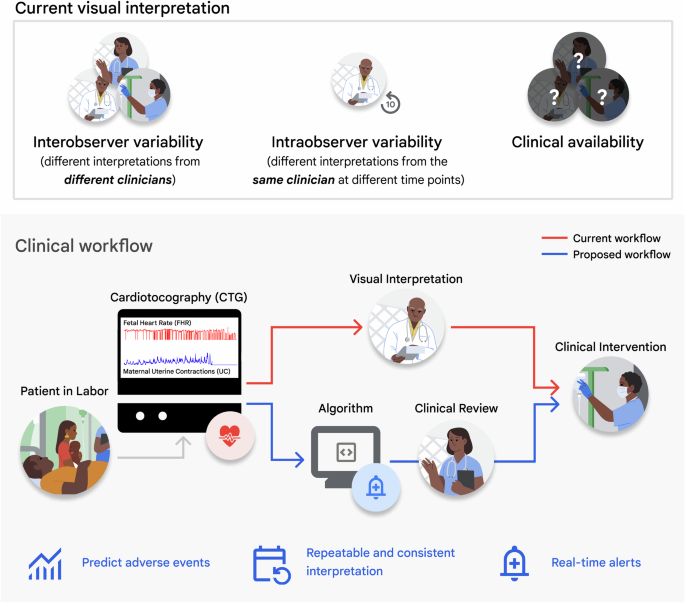
Top: Challenges of current visual CTG interpretation. Bottom: Proposed clinical use case of deep learning algorithms for CTG interpretation and assistive clinical decision-making.
Machine learning algorithms to classify abnormal CTGs from tabulated rules-based extraction of diagnostic features have shown promise for improving clinical decision support7,8,9,10,11,12. However, they reduce rich CTG signal information to a few numbers which ignore important temporal and contextual cues such as the relative timing of delivery, maternal risk factors, etc.13,14,15,16.
Current deep learning methods for CTG interpretation, which use the physiological time series data as input, rely on proxy labels for fetal well-being recorded immediately after delivery: the umbilical artery blood pH and the 1-minute Apgar score17,18,19,20,21,22,23. Umbilical cord blood pH at the time of birth, often used in high-resource medical facilities, is presently the only objective quantification for the potential occurrence of fetal hypoxia during labor. In contrast, the Apgar score at one minute from delivery is a subjective score from 0 to 10 assigned by a clinician and reflects the general health of the newborn. Apgar scores are the primary delivery outcome descriptor in low-resource settings due to their simplicity, cost-effectiveness to adopt, and the potential financial burden of umbilical blood analysis24,25.
Additionally, while CTG is widely recorded continuously in high-resource facilities, various system and implementation challenges may constrain recording to intermittent periods, which may often exclude the signal interval immediately preceding delivery26,27,28. Thus, to enable applications in such settings, machine learning-based solutions must aim to accurately detect fetal compromise at arbitrary time points before delivery to enable timely obstetric intervention26,28,29.
Our main contributions are listed as follows:
-
We highlight the feasibility of using deep learning methods to reduce the subjectivity of predicting fetal hypoxia from visual CTG interpretation.
-
We conduct evaluation studies to analyze the effect of (a) the choice of objective (pH) vs subjective (Apgar) ground truth labels, (b) the signal time interval used for training and testing, and (c) the evaluation of simulated intermittently-acquired CTG signals, on predictive performance.
-
We propose end-to-end preprocessing, data augmentation, and statistical evaluation methods to overcome challenges with this limited dataset.
-
Finally, we discuss the implications of training deep learning models for deployment in real-world use settings.
Results
Evaluating model performance by prediction task
We evaluate model performance by prediction outcome, comparing training using pH, Apgar, and the logical inclusive “OR” of the abnormal pH and Apgar criteria (LOR) ground truth labels. Our baseline method for predicting LOR, which incorporates both FHR and UC signals, achieves an area under the receiver operating characteristic curve (AUROC) of 0.68 ± 0.07, closely matching previous results (0.68 ± 0.03) obtained on a privately-owned dataset19. However, when trained and evaluated on the CTU-UHB open-source dataset, our method pipeline demonstrated higher average AUROC performance (0.68 ± 0.07) compared to the downsampling preprocessing and default convolutional neural network (CNN) architecture from the same study (0.57 ± 0.08)19. We found no statistically significant differences in sensitivity at 90% specificity between our proposed pipeline (0.27 ± 0.18) and clinician performance (0.45, 95% CI: 0.23–0.68) as reported in an observational study with 500 patients30. Additionally, there were no statistically significant differences between the baseline AUROC for Apgar prediction (0.69 ± 0.12) and LOR (0.68 ± 0.07), or between baseline pH prediction (0.62 ± 0.09) and LOR (0.68 ± 0.07). A summary of model performance is provided in Table 1, with confusion matrices for the top-performing models shown in Fig. 2h.
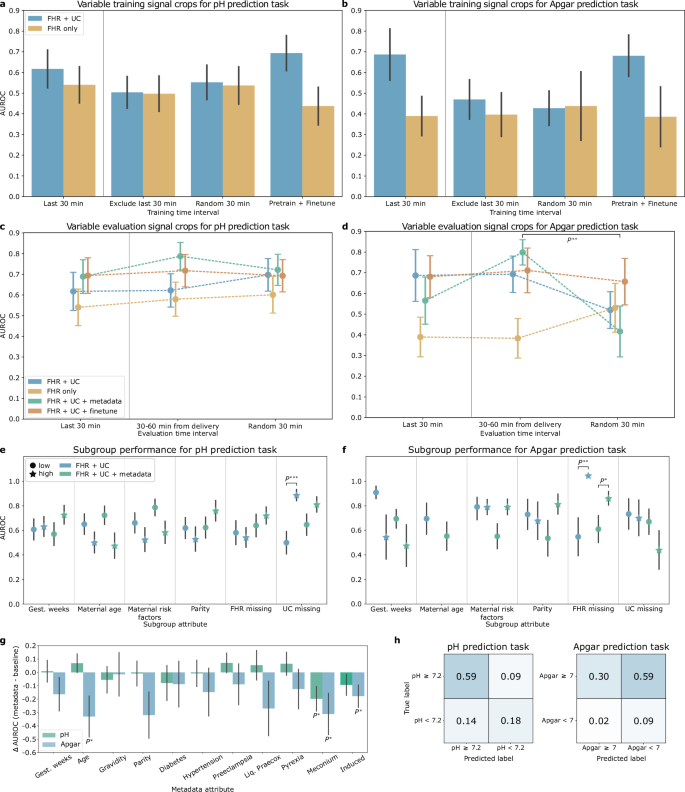
Error bars depict the standard error, computed over 1000 bootstrap samples. a AUROC for pH classification models trained on cropped signal from different time intervals. b AUROC for Apgar classification models trained on cropped signal from different time intervals. c AUROC for pH classification models evaluated on cropped signal from different time intervals. d AUROC for Apgar classification models evaluated on cropped signal from different time intervals. Markers to the left of the vertical gray line indicate the paradigm used to train and evaluate the baseline models. e Subgroup AUROC performance for the FHR + UC and FHR + UC + metadata pH classification baseline models. f Subgroup AUROC performance for the FHR + UC and FHR + UC + metadata Apgar classification baseline models. Results for the high maternal age subgroup are omitted and further details can be found in Supplementary Note 1. g Change in AUROC performance when adding a single metadata attribute to the baseline FHR + UC model input, compared to baseline. h Confusion matrix for the best-performing pH (left) and Apgar (right) prediction tasks. P*, P**, and P*** represent a p-value < 0.05, 0.01, and 0.001 respectively.
We also compare the performance obtained by deep learning and feature-based approaches for the baseline model utilizing both FHR and UC signals. For pH classification, the AUROC of the feature-based approach (0.61 ± 0.06) was not statistically different from the CNN baseline (0.62 ± 0.06). However, in the Apgar prediction task, the feature-based approach achieved a statistically significantly lower AUROC (0.35 ± 0.10 than the CNN baseline (0.69 ± 0.12).
Evaluating model performance by input signal type
We compare the performance of the deep learning model with FHR only, UC only, and FHR + UC signal input. The FHR + UC model achieved the highest AUROC performance for both pH and Apgar classification tasks, followed by UC only and then FHR only models. Excluding either of the channels also resulted in a statistically significant reduction in sensitivity at 90% specificity for both tasks.
Performance with maternal and fetal metadata
We evaluate the effect of incorporating additional clinical context via maternal and fetal metadata on predictive performance. Adding a metadata vector to the FHR + UC model input increased the performance for the pH prediction task by 0.07 points, reaching 0.69 ± 0.08, though the improvement was not statistically significant. Thus, while metadata is important, we cannot conclude that including all the provided metadata attributes had a meaningful impact on the model performance. Furthermore, adding metadata for the Apgar prediction task resulted in a non-significant decrease in FHR + UC model AUROC (0.57 ± 0.11).
We perform an ablation study to identify the significance of individual metadata component contributions to model performance. For the pH prediction task, adding an indication of meconium presence in the amniotic fluid statistically significantly decreased model AUROC (0.42 ± 0.08) compared to the baseline (0.62 ± 0.06). The addition of maternal age (0.69 ± 0.09), pre-eclampsia status (0.69 ± 0.08), premature rupture of membranes (liq. praecox) (0.67 ± 0.08), and pyrexia (0.68 ± 0.09) each contributed positively to model performance, but improvements were not statistically significant. For the Apgar classification task, the addition of metadata generally decreased performance. We observed a significant decrease in AUROC compared to the baseline FHR + UC model when including the maternal age, meconium, and induced delivery attributes. Full results are shown in Fig. 2g. The lack of significance in some of these results indicates that metadata may not be essential for predicting pH-based fetal hypoxia or 1-minute Apgar scores and might even reduce model performance in the case of Apgar. However, in standard machine learning development, even minor changes in AUROC can be important. Further studies on larger datasets and real-world prospective studies may be required to confirm the impact of metadata on predictions.
Evaluation of temporal distribution shifts during training and testing
Experimental results for training models on different time points of the CTG recording and evaluating on the last 30 min of the held-out set are shown in Fig. 2a, b. We observed no significant differences in AUROC from training on different time points and testing on the last 30 min for both pH and Apgar prediction tasks. However, Apgar prediction performance had higher variability across the different trained models compared to pH prediction performance, which was more stable. Pre-training on cropped signals before the last 30 min, then fine-tuning on the last 30 minutes achieved the highest AUROC for predicting pH using FHR + UC (0.69 ± 0.09) followed by the model trained on the last 30 min alone (0.62 ± 0.09). This agrees with clinical practice that the CTG signal recorded closest to delivery corresponds more with the recorded pH measurement. For the Apgar prediction task, pre-training and fine-tuning (0.68 ± 0.10) the model achieves a similar performance as training on the last 30 minutes alone (0.69 ± 0.12).
Experimental results for training models on the last 30 min and evaluating on different time points of the held-out set are shown in Fig. 2c, d. We observed no significant differences in the performance of the models when tested on cropped signals across different time points for the pH classification task, simulating the intermittent CTG evaluation setting. In general, pH performance remained stable across different time point evaluations. Apgar prediction performance typically had higher variability across different time points, demonstrating reduced robustness to temporal distribution shifts.
Subgroup evaluation
We evaluate subgroup-specific performance metrics to identify potential performance disparities. Figure 2e, f show the AUROC performance of the subgroup analysis for pH and Apgar prediction, respectively. We found significant differences in baseline AUROC performance between subgroups with low and high UC signal missingness with pH evaluation (low: 0.50 ± 0.09, high: 0.89 ± 0.05) and for FHR missingness subgroups with Apgar prediction (low: 0.52 ± 0.15, high: 1.00 ± 0.00).
With metadata, the performance disparities observed with pH prediction were mitigated (low: 0.65 ± 0.09, high: 0.81 ± 0.06). However, including metadata increased the AUROC performance disparities for demographic and clinical-related subgroups on this task, although none of these differences were statistically significant. This suggests that conditioning on demographic and clinical subgroup information may yield less equitable performance, though further investigation with larger sample sizes is needed to confirm the effect of metadata attributes on model performance disparities.
Discussion
Our study establishes the viability of employing deep learning for predicting fetal hypoxia from cardiotocography (CTG) tracings and the need for rigorous evaluations by choice of label, time interval, and subgroup performances. The baseline model, incorporating fetal heart rate (FHR) and uterine contraction (UC) signals, achieved performance levels consistent with prior work and clinical practice19,30. For the pH prediction task, our findings indicate that including both FHR and UC channels is imperative for accurately identifying fetal hypoxia cases. Comparative analyses with different ground truth labels revealed that objective pH measurements yield more consistent performance than subjective clinician-assigned Apgar scores, which represent more heterogeneous types of fetal compromise. Our research highlights the limitations of relying on Apgar scores as ground truth labels and encourages future work directed at predicting quantitative objective measures, such as umbilical cord blood pH. This insight is particularly pertinent for machine learning practitioners aiming to train machine learning models in medical settings lacking objective umbilical cord pH measurements24,25.
The superior performance of pH classification over Apgar classification aligns with existing literature, which predominantly focuses on binary classification of fetal hypoxia, or acidemia, using arterial cord pH thresholds between 7.05 and 7.20 for defining abnormal groups18,21,23,31. Few studies explore performance differences across pH thresholds in conjunction with annotations for severe fetal compromise. For instance, ref. 21 reported low true positive rates for detecting severe fetal compromise without acidemia using CTG time series and hypothesized that such cases represent a heterogeneous and challenging group to identify. Unlike their work, which defined severe compromise based on a composite outcome of various long-term neonatal outcomes, we focus on the 1-minute Apgar score, a heterogeneous yet immediately assessable measure of fetal compromise. By concentrating on the classification of short-term neonatal outcomes, our study lays the groundwork for future investigations into their connection with longer-term outcomes.
Furthermore, training models on the last 30 minutes of CTG recording, the time interval most closely correlated with the delivery outcome, yielded the best performance. Notably, pre-training the model on cropped signal data excluding this critical interval, followed by fine-tuning on the last 30 minutes, further improved performance. The robustness of our pH classification model to out-of-distribution time points was demonstrated by consistent performance across randomly sampled intervals within 90 minutes of delivery, simulating the intermittent CTG monitoring setting. Subgroup analyses identified performance disparities across demographic, clinical, and signal quality subgroups in the baseline model. Incorporating all available fetal and maternal metadata during training led to a non-significant increase in pH classification AUROC but worsened performance disparities within demographic and clinical subgroups. Furthermore, the inclusion of meconium presence as a clinical variable significantly decreased pH classification AUROC, while adding other clinical risk indicators (maternal age, pre-eclampsia, liq. praecox, and pyrexia) yielded non-significant positive contributions to model performance. Although these point-wise improvements in model AUROC are noteworthy, the absence of significant performance differences suggests that future studies exploring larger datasets and real-world prospective data are needed to confirm the impact of metadata on predictions.
In contrast to previous approaches8,9,10,11,12,13, we leveraged deep learning to enable end-to-end prediction of fetal hypoxia, taking into account temporal and contextual cues often overlooked by feature-based methods. The robustness of our model highlights the compatibility of our approach with intermittent CTG monitoring settings.
This study had several limitations that constrain the generalizability of our findings. First, we used CTGs from 552 patients at a single hospital in Prague, Czech Republic. To enhance the robustness of our findings, future investigations should involve a larger and more diverse dataset sourced from maternity centers worldwide, encompassing varied clinical contexts, demographics, and outcomes. Secondly, in the absence of automated CTG digitization infrastructure, the simulation of intermittent CTG use cases from facilities with digitized recordings becomes necessary32,33. Additionally, our study did not include a comparison of algorithmic performance against clinicians viewing the same dataset, prompting future research to explore different human and algorithmic use combinations. Finally, further work is needed to understand how such prediction algorithms can be optimally integrated into clinical workflows to improve neonatal outcomes.
In summary, we develop neural networks and feature-based models to interpret CTGs and propose a framework to evaluate these models. Our major findings indicate that utilizing objective pH measurements, as opposed to clinician-defined Apgar scores, results in more consistent, robust performance under temporal distribution shifts. This is especially important when transferring models to settings that only have intermittent CTG measurements. The model and evaluation framework we propose can be applied more generally to paired time-series datasets especially where sample size is limited.
Methods
Dataset description
The CTU-UHB Intrapartum Cardiotocography Database is an open-source selective collection of 552 CTGs from University Hospital Brno, Czech Republic consisting of ~50,000 minutes of recordings at term (≥37 weeks gestation)34,35. Each CTG records the fetal heart rate (FHR) and corresponding uterine contractions (UC) for up to 90 minutes before delivery. In this work, we use ‘CTGs`, ‘CTG recordings`, and ‘records` to refer to the entire patient CTG recording. In contrast, terms such as ‘input signal’, ‘signal crops’, and ‘cropped signal’ refer to the 30-minute segments of the CTG recordings used as input for the deep learning model. The data are associated with fetal outcomes, along with fetal and maternal metadata, summarized in Table 2. A correlation matrix of the associated metadata is included in Fig. 3a. This publicly available dataset is accessible at https://physionet.org/content/ctu-uhb-ctgdb/1.0.0/. According to the dataset’s documentation, all data were collected with signed informed consent and all identifying information of the participants was removed to ensure anonymity34. This retrospective study on de-identified data was reviewed and waived for further review by Advarra IRB.
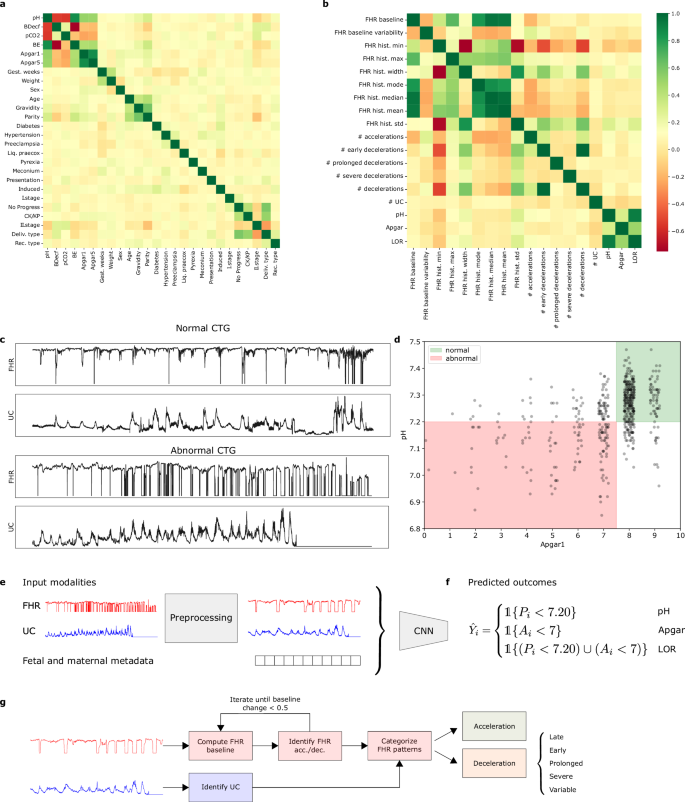
a Correlation between patient metadata attributes. b Correlation between extracted features and predicted target variables. c Top: Normal CTG recording with pH ≥ 7.20 and Apgar at 1-minute ≥ 7. Bottom: Abnormal CTG recording with pH < 7.20 and Apgar at 1 minute <7. d pH and Apgar scores for all patient CTG recordings. The green region denotes the intersection of normal recordings and the red region denotes the intersection of abnormal recordings. This figure demonstrates that there is not necessarily a direct link between pH values and Apgar scores. e Possible input modalities for the deep learning model. f Possible predicted outcomes output by the deep learning model, depending on the classification task. g Rules-based feature extraction pipeline.
We defined three outcome label categories:
where ({mathbb{1}}{cdot }) is the binary indicator function, Pi is the umbilical arterial cord blood pH, Ai is the 1-minute Apgar score, and Yi is the assigned ground truth label (abnormal = 1, normal = 0) for the ith CTG recording. The pH classification task yielded 375 normal and 177 abnormal cases, with a cut-off threshold of 7.20. This threshold was chosen for relevance to clinical cut-offs, appropriate class balance, and to enable comparison to prior work on a similar task19. The Apgar classification task yielded 484 normal and 68 abnormal cases. The LOR classification task, defined as the logical inclusive “OR” of the abnormal pH and Apgar criteria yielded 354 normal and 198 abnormal cases19. Examples of normal and abnormal CTGs are shown in Fig. 3c. Figure 3d shows the relationship between pH and Apgar scores for all patient recordings.
Preprocessing overview for the neural network methods
CTG recordings are prone to fetal and maternal movement artifacts, displacement of transducers, vaginal examinations, and maternal pushing that add noise29. Therefore, raw CTG data require preprocessing before analysis with machine learning methods since low-quality data negatively impacts the stability and convergence of learning from data36.
In our preprocessing pipeline for CTG recordings input into the neural network models, we first removed repeated missing values at the beginning and end of the recording, as missing FHR signal is non-informative for fetal status interpretation29,37. Corresponding segments of the UC signal, recorded simultaneously with the FHR signal, were also deleted38. To assess the quality of the remaining signal, we categorized missing intervals into short and long segments. Following clinical guidelines, we marked missing FHR signals longer than 15 seconds as missing values and set them to zero to maintain temporal dependencies in the data23,39. We imputed short segments of missing values using linear interpolation11,40. Both the FHR and UC signals were then smoothed to reduce the effect of noise on our classification algorithm19,29,41,42,43. We conducted data augmentation and oversampling of abnormal CTGs to address class imbalance in the dataset11,23,36,44.
To maintain consistency with the baseline method from ref. 19, we cropped recordings to 30 min segments for training and evaluation. Finally, we downsampled the recordings to 1 Hz to reduce input dimensionality, decreasing training time and hardware requirements19,21,23,29,39,42,45,46,47. This resulted in 4,315,200 minutes (n = 496 recordings), 148,800 min (n = 496 recordings), and 1,680 min (n = 56 recordings) of cropped signal for pre-training, training, and testing, respectively. A step-by-step depiction of the preprocessing pipeline is shown in Supplementary Fig. 1.
Alternative time interval preprocessing
To train models on cropped signals from different time intervals we used the following: (1) cropped signal with a sliding window of a 30 min interval and 1-minute step size that excluded the last 30 minutes of recording and (2) randomly cropped 30 min windows sampled over the entire recording. (3) We also investigated pre-training on cropped signals from (1) and then fine-tuning model parameters on the last 30 min of the recording in the training set. We partitioned the CTG recordings into identical disjoint training and testing sets for both the pre-training and fine-tuning phases to ensure that none of the patient recordings in the held-out test set were previously seen by the fine-tuned model during pre-training. The pre-training signal crops were generated by systematically cropping the available preprocessed CTG excluding the last 30 minutes of the recording using a 30 min sliding window with a 1 min stride. The number of cropped signals generated for each CTG scales with the duration of the preprocessed CTG record, so an unequal number of signals were generated for different record identifiers.
We evaluated the baseline model trained on the last 30 minutes of recording on the following varied 30 min segments in the held-out dataset: (1) the cropped signal at the 30-60 minute mark before delivery and (2) randomly cropped 30-minute signals sampled over the entire recording. These time intervals simulate the type of observed CTG recording common in intermittent CTG monitoring settings. These samples had no additive multi-scale noise and were generated in a fully deterministic fashion.
Data splitting
Due to the imbalance of classes and small patient sample size in the CTU-UHB dataset, we performed stratified data splitting to ensure similar distributions over the predicted target variable across training, validation, and test splits. Since the augmented dataset had a one-to-many correspondence between record identifiers and augmented cropped signals, we first assigned 10% of the record identifiers to a held-out test set before assigning the remaining 90% to 10-fold cross-validation (cv) splits by record identifiers. This was done to maintain a representative distribution of labels in each split and ensure no leakage of cropped signals in the training and test sets originating from the same CTG recording. The number of preprocessed CTG recordings in each training, validation, and test split for the various datasets used to train and evaluate the neural network models is summarized in Supplementary Table 2.
Neural network model architecture and training
We adapted the CTG-net neural network model proposed by ref. 19 as our base model. The CTG-net architecture takes signals of 1800 time points long (30 min downsampled at 1 Hz) as input. The input FHR and UC signals are convolved with 30-second temporal filters before a depthwise convolution is conducted to learn the relationship between FHR and UC. A final separable convolution is applied before all features are flattened and passed to fully-connected hidden layers for classification. The output of the final layer is passed through a sigmoid activation layer to yield an abnormality score. A high-level depiction of the training pipeline is shown in Fig. 3e, f. Further details regarding the architecture can be found in ref. 19.
We also ran the following experiments: (1) training with FHR or UC as input using a 1D CNN model variation and (2) adding metadata features as a vector to the input. We performed hyperparameter tuning through an architecture search to optimize the number of convolutional filters at each layer and compare our method against the default training pipeline employed by ref. 19. Model hyperparameters were optimized separately for two-channel (FHR and UC) versus 1-channel (FHR) input models. The range of values considered and the sampling functions used to conduct the model architecture and training hyperparameter search are depicted in Supplementary Table 5. The resulting optimized model architecture parameters and fixed hyperparameter values are shown in Supplementary Table 6. Out of 500 random hyperparameter configurations, the best hyperparameters were selected based on the highest validation AUROC averaged over 10 cross-validation folds. The same procedure was used to select the model from the pre-training phase to use for downstream fine-tuning.
All neural networks were trained on an NVIDIA V100 GPU in TensorFlow (https://www.tensorflow.org) using the Keras API (https://keras.io). We used Adam as the optimizer, initialized model weights and optimizer state with a fixed random seed, and trained each model for 300 epochs48. We perform channel-specific maximum absolute value scaling on time series and metadata normalization on tabular features before input into the neural network.
Rules-based classification with XGBoost
To compare the performance of the neural network model to conventional machine learning algorithms, we implemented a feature extraction pipeline to extract FHR and UC features from the time series signal according to the current maternal and fetal medicine practices of the International Federation of Gynecology and Obstetrics (FIGO)37. The following features were extracted: uterine contractions, FHR baseline, baseline variability, accelerations, decelerations, variable decelerations, severe decelerations, late decelerations, prolonged decelerations, and the width, minimum, maximum, median, mean, mode, and standard deviation values of the FHR histogram. Figure 3b shows the correlation matrix for all extracted features and predicted target variables. Figure 3g depicts the feature extraction pipeline.
Preprocessing steps up to and including the imputation of missing values were applied to generate a separate dataset to use with the feature extraction algorithm. Following the imputation of segments shorter than 15 seconds, the FHR and UC signals were smoothed with a rolling window of 120 time points (30 seconds), and missing segments longer than 15 seconds were imputed using linear interpolation. The same small-scale edge clipping and additive noise data augmentation process was applied to the smoothed signal.
The uterine contractions were identified using a peak-finding algorithm. The initial FHR baseline was computed over a 10-minute window and iteratively updated as accelerations and decelerations were identified and removed from the baseline calculation37. This process was repeated until the magnitude of the FHR baseline change was less than 0.5. Finally, the baseline variability, FHR accelerations, and decelerations were detected according to the estimated FHR baseline. We implemented the feature extraction algorithm in Python and validated the extracted features for several CTG recordings with a practicing OBGYN on our team.
We trained an XGBoost classifier, typically used in CTG applications8, on the extracted FHR and UC features for the pH and Apgar classification tasks using the same data splits used to train the neural network models. We used 1,000 estimators with a maximum depth of 2, a class-corrected weighted loss, 30 early stopping rounds, and AUROC as the evaluation metric.
Evaluation
The performance of the neural network and XGBoost models was evaluated using the area under the receiver operating characteristic curve (AUROC). We also reported the sensitivity at a fixed specificity threshold of 90% for comparison with clinical performance.
A two-tailed Welch’s t-test was used to compare the average AUROC computed over 1000 bootstrapped samples for the various approaches. In cases where we compared performance on a fixed test set, a paired t-test was used. We also evaluated performance disparities across subgroups for both pH and Apgar prediction tasks. Subgroup variables included demographic and clinical attributes (gestational age of the fetus, maternal age, maternal risk factors, parity)49,50,51 as well as signal quality descriptors for each of the FHR and UC channels. A comprehensive description of the cut-offs and formulas used to define binary subgroup variables is found in Supplementary Table 3. Signal quality metrics were computed after the cropping step in the preprocessing pipeline and are summarized for each time interval in Supplementary Table 4.





Responses