A latent Axin2+/Scx+ progenitor pool is the central organizer of tendon healing

Introduction
Stem and progenitor cells rely on signaling interactions with their niche environment to maintain their state and regulate tissue maintenance and repair1. These signals can originate from neighboring cells, the extracellular matrix (ECM), daughter cells, or from the cells themselves2,3,4,5,6. Dense matrix tissues such as tendons present a unique constraint for progenitor cell regulation as there is a low cell density, limited turnover, and few cell–cell contacts. Yet despite these conditions, tendon cells can mobilize after injury to proliferate and repair the damaged tissue7,8,9. Currently, the mechanisms underlying such regulation and the molecular identity of resident stem and progenitor cell populations are poorly defined. Identifying central pathways and cell types capable of repairing tendon tissue in vivo would have major therapeutic implications as tendon injuries are a common clinical problem, accounting for up to 50% of all sports and work-related injuries10.
The tendon’s ECM consists of highly ordered type I collagen fibrils, which serve an important function in force transmission from the muscle to the bone. The hypocellular tendon is traditionally considered to contain two main populations of cells defined by their location and morphology: peritendinous cells that surround the tendon in a sheath-like structure and elongated tenocytes with their long cellular projections within the main tendon body11. Tenocytes reside within the ECM of the main tendon body and express the tendon transcription factor, Scleraxis (Scx) which can be detected by the Scx transgene (Scx)-GFP, along with tendon matrix components including, Collagen1a2 (Col1a2) and Tenomodulin (Tnmd)12. Recent studies have revealed more tendon cell heterogeneity9,13, than was previously appreciated but the function of distinct tendon cell subsets remains unclear. Cells termed tendon-derived stem/progenitor cells (TDSPCs) were identified ex vivo through their characterization by surface marker expression, expansion, and ability to undergo serial transplantation into mice14, but their relationship to resident tendon cell populations in vivo is unknown. Previous analysis of radioisotope incorporation in human adult tendons has shown limited bulk tissue turnover after the age of adolescence15, making it unclear what role tendon stem or progenitor cells may have in maintaining adult tendon tissue. However, analysis of diseased tendon samples revealed increased tissue turnover rates16. In mice, low levels of proliferation of Scx–GFP+ cells have been detected throughout adulthood17, and upon tendon injury, a robust cellular response occurs, which together, could be suggestive of stem or progenitor cell activity. In identifying the origins of these contributing cell populations, previous lineage tracing studies have shown the contribution of Sma+ and Tppp3+ cells from the surrounding sheath to tendon healing7,8,9, pointing to the presence of tendon stem or progenitor cells in the tendon sheath. Other studies have shown major contributions to adult tendon healing from Scx-lineage cells18,19. However, it is unclear what mechanisms maintain the ability of the cells to respond to injury and initiate their migration and proliferation in healing.
Wnt signaling plays a crucial role in regulating embryonic development and stem/progenitor cell behavior in various tissues and organs throughout the body. Wnt signaling is commonly classified as canonical, which acts through β-catenin nuclear localization and transcriptional regulation, or non-canonical, which signals through GTPases or kinases that lead to transcriptional changes. Whereas non-canonical Wnt signaling is linked to changes in cell polarity and migration, canonical signaling has been shown to regulate stem cell self-renewal, proliferation, and regeneration20,21. Several studies have shown the importance of canonical Wnt signaling in both marking and regulating stem cells in the hair follicle, intestinal epithelium, and mammary gland22,23,24. Particularly in the skeleton, Wnt has been shown to regulate the conversion to osteoblast precursor cells during bone regeneration25. A recent study showed that cells marked by Axin2, a direct transcriptional target and negative regulator of canonical Wnt signaling, contributed to neonatal tendon healing26. However, the function of Axin2+ cells and Wnt signaling has not been explored in the adult tendon.
Here, we describe a latent, yet injury-responsive, Axin2+ tendon cell population that is regulated by their own Wnt secretion. After a tendon injury, the Axin2+ cells mount a robust injury response by down-regulating Scx-GFP, migrating to the wounded area, proliferating, and adopting a rounded cell morphology. Expanded and transplanted Axin2+ cells differentiate into tenocytes in an injured host tendon with a subset of lineage-marked cells expressing Axin2, demonstrating progenitor cell activity. In adults at homeostasis, the Axin2+ cells express Scx–GFP and divide more slowly than their non-Axin2 counterparts. However, ex vivo, they readily expand, display multi-lineage differentiation potential, and are enriched for the same markers found in TDSPCs. Single-cell RNA-sequencing analysis of adult tendons in homeostasis and healing reveals complex heterogeneity in tendon cell populations and underscores the central role of Axin2+/Scx+ cells in healing. During the healing process, Axin2+ cells are enriched for Wnt ligands and Wnt pathway components and at early healing stages do not express Scx-GFP. Later in healing, Axin2+ cells differentiate into tenocytes expressing characteristic tendon genes, including Scx-GFP, with elongated cellular morphology. We show that Scx+ and Axin2+ tendon cells self-regulate their own Wnt ligand expression, Axin2+ identity, and activation upon injury via autocrine Wnt signaling. Furthermore, loss of Wnt signals specifically from Axin2+/Scx+ cells severely compromises tendon healing, underscoring their key role in initiating the healing response. Complementary lineage tracing and loss of function analysis targeting Scx+ tenocytes demonstrate similar results, supporting the notion that this activity resides in Axin2+/Scx+ cells. Thus, Axin2+/Scx+ cells represent a subpopulation of quiescent tendon cells that self-regulate their maintenance and mobilization upon injury.
Results
Axin2
TdTom cells are major contributors to tendon healing
The Wnt responsive lineage-tracing mouse line, Axin2CreERT2 27,28, was found to label a subset of cells in adult tendons using either the ROSALSLmTmG or ROSALSLTdTomato (abbreviated hereafter as Axin2TdTom) reporter lines (Supplementary Figs. 1 and 2A, B). As there are only a few markers of tendon cell heterogeneity and since Axin2 marks stem and progenitor cells in other tissues29,30,31, we sought to further define this cell population in the tendon. Multiphoton microscopy allowed us to simultaneously visualize collagen organization and density via second harmonic generation imaging (SHG). Multiphoton imaging of adult tendons in Axin2TdTom mice revealed TdTom+ cells with elongated tenocyte morphology embedded in the collagen matrix, in the peritendinous tissue or sheath, as well as in surrounding non-tendinous regions predominantly along the vasculature, likely pericytes (Supplementary Fig. 1A–E’ and Supplementary Movies 1 and 2). We next compared Axin2TdTom-labeling with the well-known tendon cell labeling line, ScxCreERT2; ROSALSLTdTomato mice (abbreviated as ScxTdTom)32. ScxTdTom cells were found within the main tendon body, referred to as the fascicles in larger vertebrates33 and a smaller subset in peritendinous regions. Quantification of TdTom-stained cryosections showed labeling in 10.16 ± 2.56% of fascicular tenocytes, 24.30 ± 3.22% of peritendinous cells, and 18.55 ± 2.72% of surrounding non-tendinous cells of Axin2TdTom limbs (Supplementary Fig. 1F, G), and 31.48 ± 4.97% of fascicular tenocytes, 8.05 ± 3.57% of peritendinous cells, and 0.66 ± 0.3% of surrounding non-tendinous cells of ScxTdTom limbs (Supplementary Fig. 1A, H–L and Supplementary Movies 3 and 4).
To determine if the Axin2TdTom cells co-express Scx, we examined Axin2TdTom cells for expression of Scx–GFP by flow cytometry and multiphoton microscopy to verify that Axin2TdTom labels adult tendon cells. After treating the mice with Tamoxifen (TAM) at postnatal day (P) 60, we found that a subset of Scx-GFP cells were Axin2TdTom-positive. Double Scx-GFP+/Axin2TdTom cells were observed in the main tendon body, entrenched between highly SHG-positive collagen fibers at P80 and comprised approximately 10% of the total analyzed cell population (Fig. 1A–D, Supplementary Fig. 2C, and Supplementary Movie 5). Therefore, Axin2TdTom marks a subset of Scx-GFP cells in the adult tendon during homeostasis.
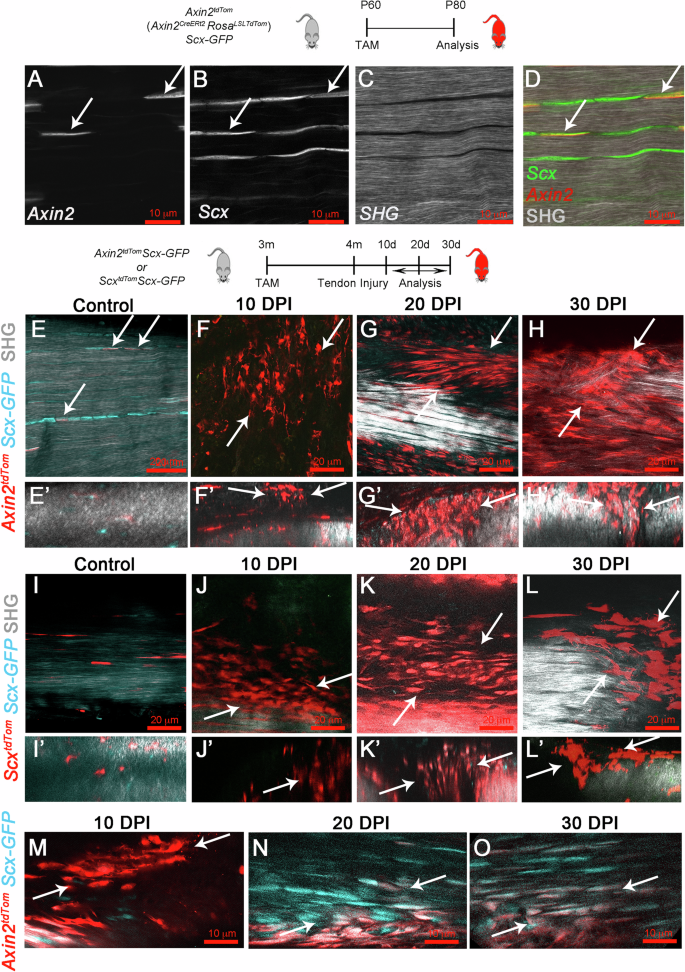
A–D Sagittal view of Axin2TdTom cells co-expressing Scx-GFP (arrows) in the main Achilles tendon body (n = 20 Achilles tendons examined) (SHG). Cartoon indicates mouse genotype and TAM was given at P60 with imaging at P80. E–O Mice of the indicated genotype were given TAM at 3 months of age and Achilles tendon injury was performed at 4 months. Two-photon microscopy images were taken of contralateral uninjured Achilles tendons of Axin2TdTom; Scx-GFP mice (E, E’) and of injured tendons at 10 dpi, 20 dpi, and 30 dpi (F–H’). Two-photon fluorescent and SHG images of contralateral uninjured (I, I’) and injured Achilles tendons from ScxTdTom; Scx-GFP mice at 10 dpi, 20 dpi, and 30 dpi (J–L’). Two-Photon images show Axin2TdTom cells at the injury site with more rounded morphology at 10 dpi (M) and co-expressing Scx-GFP with elongated morphology at 20 dpi and 30 dpi (M–O). E–O Circle indicates the site of injury; n > 6 mice were examined per time point and genotype. E–O are sagittal views; E’–L’ are transverse views digitally resliced from sagittal sections of 0.4 μm.
To understand if Axin2TdTom cells are involved in the tendon healing response, we performed genetic lineage tracing of Axin2TdTom cells using a mouse partial acute excisional injury model of the Achilles tendon. We treated 3-month-old mice with TAM to lineage trace Axin2TdTom cells and at 4 months, performed a biopsy punch injury in the midbody of the Achilles tendon (Supplementary Fig. 2D, E). We examined cell lineage and collagen organization and density in healing tendons using 2-photon microscopy and SHG imaging. At 10 days post-injury (dpi), the wound site was infiltrated with Axin2-lineage cells, and the SHG signal was impaired compared with uninjured contralateral control tendons (Fig. 1B, E–F’). Axin2-lineage cells did not express Scx-GFP at 10 dpi (Fig. 1F, F’, M), suggesting Scx-GFP is downregulated upon injury. At 20 dpi and 30 dpi, the Axin2TdTom cells co-expressed Scx-GFP and their cell morphology changed from round to elongated, indicative of tenocyte differentiation (Fig. 1G, H’, N, and O and Supplementary Movies 6–8). Importantly, we found that the behavior of Axin2-lineage cells in response to injury mirrored that of Scx-lineage tendon cells, which have been shown to infiltrate healing sites in injuries in other tendons34. Lineage tracing of Scx-labeled tendon cells after injury revealed that similar to Axin2TdTom cells, ScxTdTom cells were observed throughout the healing site and were negative for Scx-GFP at 10 dpi (Fig. 1I, J”,). By 20 dpi, ScxTdTom cells became elongated in appearance and expressed Scx-GFP (Fig. 1K, L”).To determine if Axin2TdTom cells proliferate after injury, BrdU was administered continuously during healing. We found that Axin2TdTom cells incorporate BrdU at the injury site significantly more than the contralateral uninjured tendons (Supplementary Fig. 2F–H). Quantification of cells showed that Axin2TdTom cells comprise a major portion of the total proliferating cells at 20 dpi and of the total cells in the healing region per defined area at 10 dpi, 20 dpi, and 30 dpi (Supplementary Fig. 2I, J). Together, these data show that Axin2TdTom cells are the major cell population that infiltrates, proliferates, and differentiates into tenocytes after injury.
Adult Axin2 cells proliferate less frequently since birth than non-Axin2
TdTom cells
To understand the formation and activity of Axin2TdTom cells in adult vs growing tendons, first, we examined Axin2 cells and Wnt signaling from stages of postnatal tendon growth to physiological homeostasis. Our previous work showed that a significant decrease in tendon cell proliferation occurs after P21. Therefore, TAM treatment was performed at the neonatal stage P2, when the tendon is growing longitudinally and has significant cell proliferation, and at P60 during homeostasis, when longitudinal tendon growth and cell proliferation have slowed17. As there is no published methodology to prospectively isolate tendon cells, we devised a negative sorting strategy to remove blood (CD45+) and endothelial (CD31+) cells. Using flow cytometry and our negative sorting strategy, we found that TAM treatment at P2 resulted in 28 ± 6% of the total cells being labeled by Axin2TdTom at P80 (Fig. 2A, C and Supplementary Fig. 3A) whereas Tam at P60 showed that 10 ± 2% of the cells were labeled by Axin2TdTom at P80 (Fig. 2B, C and Supplementary Fig. 3B). These results could indicate that the number of Axin2TdTom cells in the tendon decreases with age or that early Axin2TdTom cells at P2 proliferate and expand during growth. To explore this possibility further, we found decreased qualitative expression of Axin2 by single-molecule fluorescent in situ hybridization (smFISH) at P28 compared to P0 (Supplementary Fig. 3C, D). In addition, we observed decreased expression of Wnt ligands and associated pathway genes and less accessible associated genomic regions over time upon integrated analysis of RNA-seq and ATAC-seq datasets of tendon cells from P0 to P35 (Supplementary Fig. 3E, F). Together, this analysis suggests that Wnt signaling decreases as the tendon shifts from periods of growth (P0–P21) to physiological homeostasis (>P35)17 and that the early P2 labeled Axin2TdTom cells may proliferate in growth during periods of increased Wnt signaling.
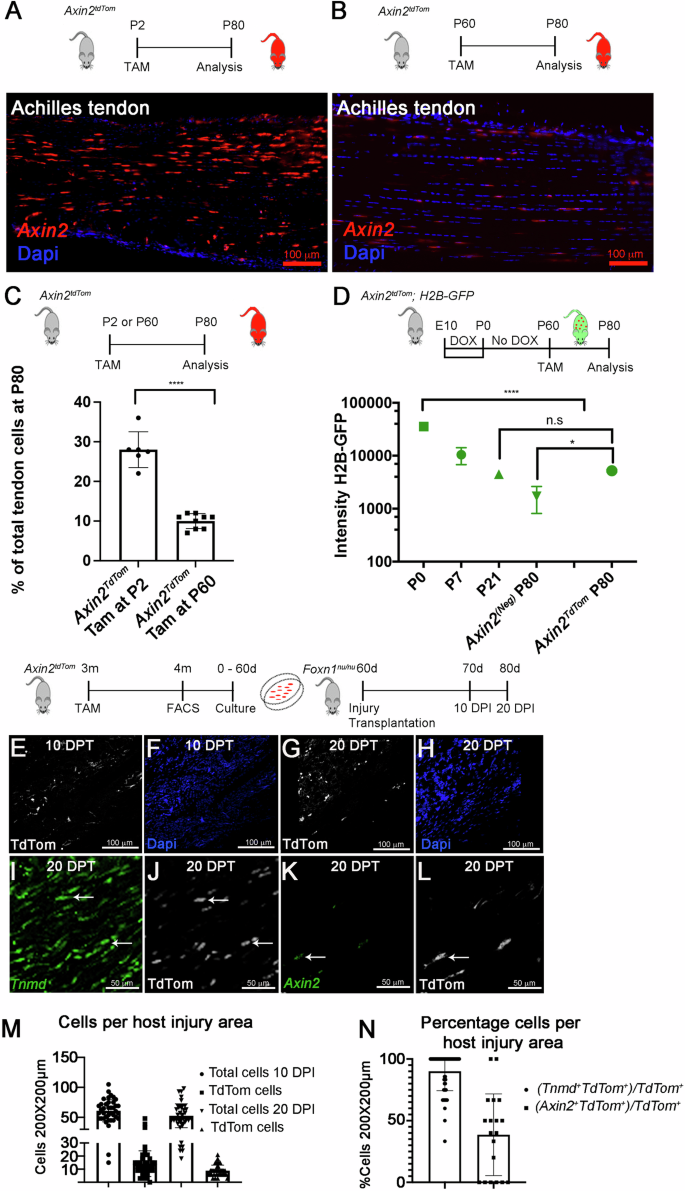
A–C TAM was given to Axin2TdTom mice at (P2) or adults (P60). Tissue was collected and analyzed at P80. Increased numbers of Axin2TdTom cells are observed upon TAM at P2 (A) compared with P60 (B) in P80 sagittal Achilles tendon sections. C Flow cytometry at P80 shows a greater percentage of Axin2TdTom cells per total tendon cells when TAM labeling occurs at P2 vs P60 (P2 cohort, n = 6 mice; Adult/P60 cohort, n = 8 mice, two-tailed unpaired T-test; ****p < 0.0001). D The DOX-inducible H2B-GFP system was used to evaluate Axin2TdTom cell proliferation from birth. DOX was administered to timed pregnant females from Embryonic day 10 (E) to birth. At birth, DOX was removed. For Axin2TdTom labeling, TAM was given at P60 and cells were analyzed at P80. CD34+/CD45+ cells were removed to enrich for tendon cells. Axin2TdTom and Axin2(Neg) cells from the same mouse were compared. Axin2TdTom cells had significantly higher H2B-GFP intensity compared to Axin2(Neg) cells at P80 (n = 4 mice/time point, one-way ANOVA, F = 148; n.s. not significant; *p < 0.05) (D). E–H Achilles tendons from 4-month-old mice with TAM at 3 months were collected and 100–500 Axin2TdTom (CD31−/CD45−/TER119−) cells/mouse were isolated by FACS. Cells from each mouse (n = 5) were expanded separately in-vitro for 60 days and a small punch of alginate-based hydrogel cell constructs were transplanted into injured tendons of immunodeficient mice (Foxn1nu/nu). The injury site was analyzed at 10 days and 20 days after injury/transplantation (DPT) (n = 9 mice, 18 injured/transplanted tendons). I–L smFISH for Tnmd and Axin2 with co-immunofluorescence staining for TdTom shows co-expression in transplanted Axin2TdTom cells at 20 DPT in sagittal Achilles tendon sections (arrowheads; n = 9 mice, 18 tendons, quantification of at least three sections/mouse). M Quantification of total cells (Dapi+) or TdTom+ cells per 200 × 200 μm area in the host injury site at 10 and 20 DPT. N Quantification of smFISH and immunostaining shows the percentage of Tnmd+ TdTom+ or Axin2+ TdTom+ cells per total TdTom+ cells in the injury area. Bars show S.D. (quantification was made using at least three locations in each section in at least three sections/tendon).
To define the proliferative activity of the adult labeled Axin2TdTom cells, we used the doxycycline (DOX) inducible histone 2B-green fluorescent protein (H2B-GFP) reporter mouse model (Col1a1:tetO-H2B-GFP;ROSA-rtTA). This system is used to quantify cell proliferation and identify slowly cycling label-retaining cell populations based on the stability and dilution of the H2B-GFP protein in each cell35,36. We activated H2B-GFP expression by administering DOX from E10.5 to P0 to timed-pregnant females. We removed DOX at birth and allowed the cells to dilute their label based on their proliferation rates as previously reported17. After administering TAM at P60, we compared Axin2TdTom to Axin2(Neg) (non-Axin2TdTom) cells from the same mouse at P80 and used GFP beads to calibrate measurements of GFP intensity and decay between experiments. We were unable to use Scx-GFP to select for tendon cells as it would interfere with H2B-GFP measurements. Therefore, to enrich the sample for tenocytes, we examined H2B-GFP presence and intensity after excluding CD31+ endothelial and CD45+ blood cells. This allowed us to determine how P60-labeled Axin2TdTom tendon cells replicated since birth in comparison to non-Axin2TdTom cells. Surprisingly, we found that almost all Axin2TdTom cells were H2B-GFP+ (95% ± 3%; n = 4 mice) compared with only 46.1% ± 16.5% Axin2(Neg) cells being H2B-GFP+at P80. Axin2TdTom cells also had significantly higher intensity than that of Axin2(Neg) cells with 5200 ± 300 vs 1716 ± 904 fluorescence value at P80 (Fig. 2D). Taken together, this indicates that the Axin2TdTom cells at P60 are derived from a cell population that divided less frequently since birth than the Axin2(Neg) cells, and suggests they are a distinct quiescent cell subset.
Axin2
TdTom cells differentiate and self-renew in a transplantation assay
As latency or quiescence is a characteristic of other tissue-resident stem or progenitor cells (reviewed in ref. 37), we next sought to test the potential of Axin2TdTom cells by examining their capacity to proliferate, differentiate, and self-renew using a cell transplantation assay into injured host tendons. After administering TAM to label Axin2TdTom cells at 3 months, approximately 100–500 Axin2TdTom cells were isolated from Achilles tendons at 4 months and cells from individual mice were cultured separately. After expanding the cells for 60 days, approximately 4 million Axin2TdTom cells per mouse were harvested, embedded in alginate gels, and biopsy punch gel constructs were transplanted into excisional defect injuries in the Achilles tendons of Foxn1nu/nu mice38. After 10 and 20 days, we examined the site of transplantation and injury for the presence of Axin2TdTom cells by immunostaining for TdTomato (Fig. 2E–H, J, and L and Supplementary Movie 9). After quantifying the number of TdTom+ and total cells per area at the injury site (Fig. 2M, N), we observed similar numbers of total cells per area at 10 and 20 days post injury and transplantation with approximately 21% ± 9% of them being TdTom+ (Fig. 2M). To determine if the expanded Axin2TdTom cells differentiated and/or retained Axin2+ identity, we performed smFISH for Tnmd, an ECM component of differentiating tendon cells, and Axin2 and co-immunostained for TdTom. We found that 90% ± 2% (standard error of means (SEM)) of the TdTom+ transplanted cells also expressed Tnmd, and 38% ± 7% (SEM) expressed Axin2 (Fig. 2N). Since Axin2TdTom cells demonstrate the ability to differentiate into Tnmd-expressing tenocytes and retain their Axin2+ state after significant expansion, we conclude that this latent tendon cell population may also have self-renewal potential.
Axin2
TdTom cells readily expand, are enriched for stem/progenitor cell markers, and undergo multilineage differentiation in vitro
As Axin2TdTom cells show potent expansion capabilities following injury and upon transplantation in vivo, we sought to further define Axin2TdTom cells using in vitro assays. We TAM-treated 3-month-old mice, harvested and isolated Axin2TdTom and Axin2(Neg) cells by flow cytometry at 4 months (isolation strategy shown in Supplementary Fig. 4), and co-cultured them for 10 days. In culture, the Axin2TdTom cells changed morphology, transitioning from thin spindle-shaped cells with long cytoplasmic extensions to more rounded cells (Fig. 3A–C). Notably, Axin2TdTom cells comprised 9% ± 2% of the cell population prior to plating, and by day 10 of culture they were 47% ± 5% of the cells (Fig. 3D, E), which represents an almost five-fold increase from their abundance in tendons. To compare the gene expression of Axin2TdTom and Axin2(Neg) cells, the cells were harvested as described but plated separately on day 0 of culture. After 10 days, RT-qPCR analysis revealed that Axin2TdTom cells had increased levels of Axin2 relative to Axin2(Neg) cells (Fig. 3F), suggestive of a persistent response to canonical Wnt signaling. We also observed greater relative expression of the tendon genes, Scx, Mohawk (Mkx), and Collagen 1a1, and of Ki67 (Fig. 3F), consistent with their tendon identity and ability to readily expand in vitro. As TDSPCs have primarily been characterized ex vivo using cell culture and transplantation assays14, the identity of the resident cell population has been elusive. To test if the Axin2TdTom cells are enriched for markers of previously described TDSPCs, we analyzed the surface marker expression of CD44, CD90.2, and Sca-1 comparing Axin2TdTom and Axin2(Neg) cells by flow cytometry after 10 days in culture, prior to passaging. We found that Axin2TdTom cells had a significantly higher percentage of CD44, CD90.2, Sca-1, and double positive CD90.2/CD44 expressing cells compared with Axin2(Neg) cells (Fig. 3G–K). The Axin2TdTom cells also were negative for CD31 and CD45 at day 10 in culture (Supplementary Fig. 4A). Together, these data show that a higher percentage of Axin2TdTom cells express the surface markers that were previously characterized for TDSPCs and are suggestive that Axin2TdTom cells could have been responsible for the activities of TDSPCs14. As the presence of Axin2 transcripts suggests the cells are actively experiencing Wnt signaling, we next tested if Wnt signaling is necessary for the proliferation and gene expression of Axin2TdTom cells. We added Wnt974, a Porcupine inhibitor, which blocks Wnt secretion, to isolated and cultured Axin2TdTom cells, and administered BrdU. Compared to DMSO treated controls, we observed decreased BrdU-incorporation and decreased Axin2 and Scx expression by RT-qPCR in Axin2TdTom cells (Fig. 3L and Supplementary Fig. 4B, C), suggesting that Wnt signaling is important for proliferation and maintenance of Scx expression in vitro. Finally, we examined the ability of Axin2TdTom cells to undergo multilineage differentiation in vitro by culturing the cells in osteogenic, adipogenic, and chondrogenic induction media. To perform these assays, we isolated and expanded Axin2TdTom cells for 60 days to obtain sufficient cell numbers. Notably, Axin2(Neg) cells did not readily expand to sufficient numbers in culture so multilineage differentiation assays were not performed. Axin2TdTom cells from individual mice were cultured separately in induction media, stained, and the number of differentiated (positively stained cells) per total cells in a defined area per well was quantified. We observed Alizarin red, Oil red O, and Alcian blue stained Axin2TdTom cells, but at differing frequencies with a higher ratio of osteogenic differentiation (Fig. 3M and Supplementary Fig. 4D). In addition, treatment with Wnt974 significantly increased the ratio of Alcian blue-stained cells (Fig. 3M), suggesting that Wnt signaling antagonizes differentiation towards the chondrogenic lineage. Together, our results show that the Axin2TdTom cells in vitro can readily expand, better retain tendon identity, are enriched for markers of TDSPCs, and have multilineage differentiation potential.
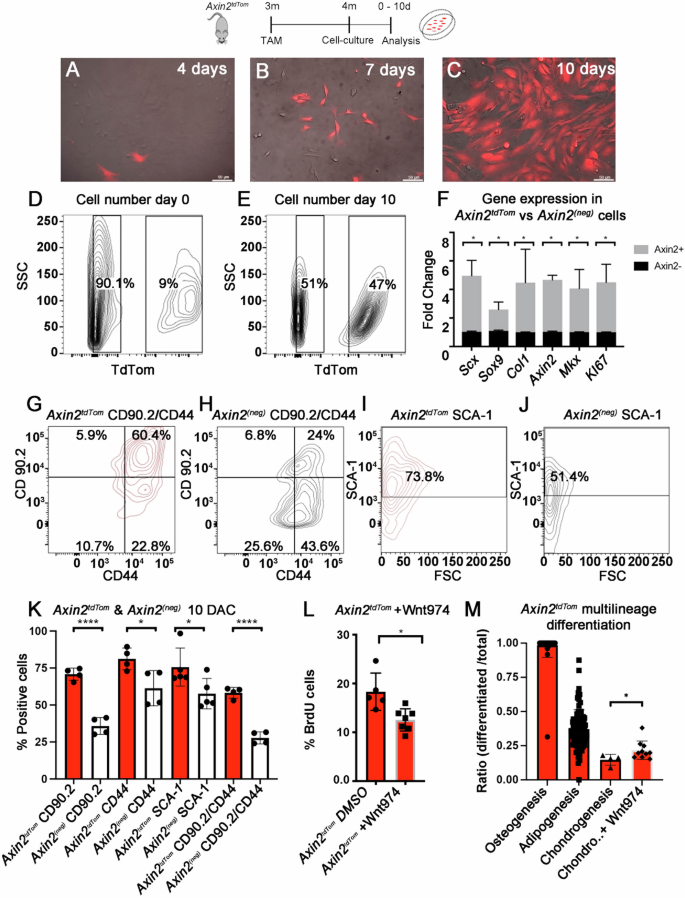
A–M TAM was given to 3-month-old mice and at 4 months, Axin2TdTom cells were extracted from the limb tendons and analyzed or plated in cell culture. A–C Images are focused on Axin2TdTom cells to show changing cell morphology at 4 days, 7 days, and 10 days in culture. D, E Representative flow cytometry plots showing Axin2TdTom and Axin2(Neg) cells prior to (0 days) and at 10 days in culture. F Axin2TdTom cells express higher relative amounts of Scx, Sox9, Col1a2, Axin2, Mkx and mKI67 transcripts by RT-qPCR relative to Axin2(Neg) cells after 10 days in culture (n = 3 mice; Multiple T-test with two-stage step-up method of Benjamini, Krieger and Yekutieli; *p < 0.05). Flow cytometry histograms show Axin2TdTom cells are enriched for CD44/CD90.2 double positive (G) vs Axin2(Neg) cells at 10 days of culture (H). I, J A higher percentage of Axin2TdTom cells express Sca-1 compared with Axin2(Neg) cells at day 10 in culture. K Quantification of CD 90.2, CD 44, Sca-1, and double-positive CD90.2/CD44 Axin2TdTom cells vs Axin2(Neg) cells shows the cells are significantly enriched for the markers (n > 4 mice per experimental group; Multiple T-test with Welch correction *p < 0.05, ****p < 0.0001). L The Porcupine and Wnt secretion inhibitor Wnt 974 or vehicle (DMSO) were added to a final concentration of 2.5 μM to cultured Axin2TdTom cells. BrdU was added to both conditions for 5 days and cells were analyzed for TdTom expression and BrdU incorporation (n ≥ 5 with each n representing cells from an individual mouse per condition were analyzed; unpaired T-test with Welch’s correction *p < 0.05). M Axin2TdTom cells from 4-month-old mice were cultured and expanded separately for 60 days. After obtaining sufficient Axin2TdTom cell numbers, adipogenic, osteogenic, and chondrogenic assays were performed and cells were stained (examples shown in Supplementary Fig. 4). Stained vs total cells were quantified per 200 × 200 μm area with at least four measurements per well, in at least three wells per treatment, and each well-represented cells from a different mouse (n = 5). Addition of Wnt974 yielded a higher percentile of chondrogenic cells (T-test with Welch’s correction *p < 0.05; each dot represents a 200 × 200 μm quantification of cells).
A single cell atlas of the adult tendon reveals activation of Scx+/Axin2
+ cells during healing
We next sought to better define Axin2TdTom cells in the context of all the cells in the tendon. Since Axin2TdTom cells and ScxTdTom cells are both found to infiltrate the injury site (Fig. 1F–L’ and Supplementary Fig. 2J), we believe the target cells overlap the original populations that are labeled by Axin2CreERT2 and ScxCreERT2 in our lineage tracing strategy. To better characterize Axin2TdTom cells as Scx-GFP+ tendon cells, we performed single-cell RNA-sequencing on 4-month-old Achilles tendons during homeostasis and at 10 dpi. Our analysis identified distinct tendon and connective tissue cell clusters, as well as other cell types, including Schwann cells, endothelial cells, skeletal muscle, and pericytes (Fig. 4A). Blood cells were present but removed from subsequent analysis. Sub-clustering of tendon and connective tissue cells revealed distinct populations that we characterized based on the enrichment of specific markers (Supplementary Table 1). The clusters include midbody tenocyte (MBT) (e.g., Scx+, Tnmd+, Thbs4+, and Fmod+), the myotendinous junction (MTJ)-associated tenocyte (e.g., Col22a1+, Chodl, and Scx+)39, osteotendinous or enthesis-associated (Ent) tenocyte (e.g., Pthlh+, Inhbb+, and Scx+)40, and osteoblast-like cells (Ost; e.g., Sox9+, Lum+, Osr2+, Smoc2+, Gli1+, and Scx−)41,42,43 in addition to four tendon fibroblast clusters (TF1–4). We did not detect Scx in TF1–4 (Fig. 4B, C), but these clusters were identified as tendon or tendon-associated by their enriched expression of Col1a1, Col3a1, and other ECM components. Two additional clusters were named injury-responsive cell states (IRC-1 and IRC-2), as they were found in homeostasis and expanded significantly upon injury.
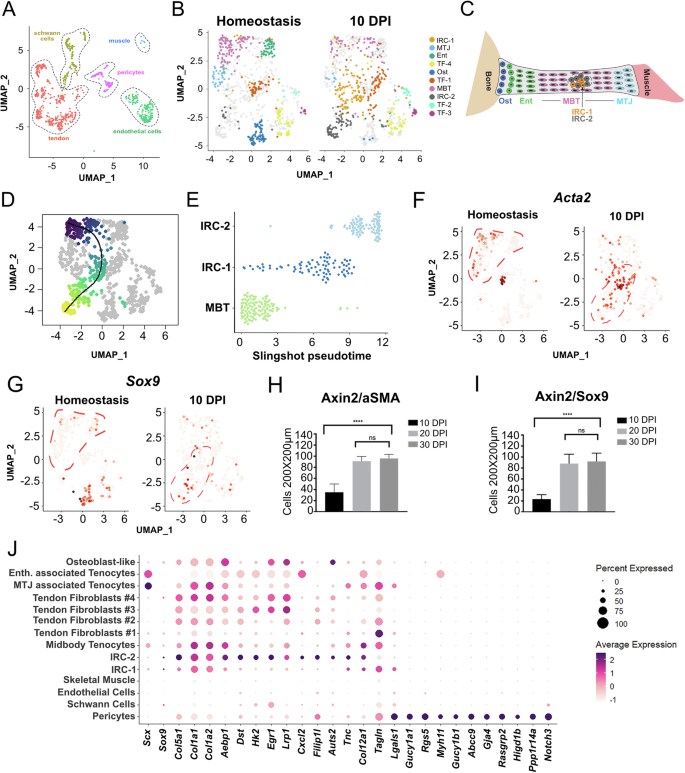
A UMAP representation of single cell RNA-seq data from Achilles tendons showing total cells excluding blood cell clusters. B UMAP of sub-clustered tendon cells in homeostasis and 10 dpi. For homeostasis, Achilles tendons from 4 month-old Axin2TdTom mice (n = 4, 8 tendons) with Tam at 3 months. For 10 dpi, three Achilles tendons were harvested (n = 3 mice) after tendon injury at 4 months of age. Ten tendon clusters were identified: MBT, MTJ, Ent, Ost, TF1–, IRC-1, and IRC-2. C A cartoon model depicting detected clusters in the tendon. D, E Slingshot pseudotemporal reconstruction analysis inferred trajectory from the MBT to IRC-1 and IRC-2 clusters. UMAP feature plots for Acta2 (F) and Sox9 (G) are shown for homeostasis and 10 dpi. Tenocyte clusters (MBT, MTJ, and Ent) are outlined in red dotted lines for homeostasis, and injury-responsive cell states (IRC-1 and IRC-2) are outlined for 10 dpi. Acta2 and Sox9 are expressed in cells in IRC-1 and IRC-2, respectively, at 10 dpi. H, I Quantification of cells co-expressing Sox9 or αSMA with Axin2TdTom per 200 × 200 μm square area of the injury site at 10 dpi, 20 dpi, and 30 dpi. Quantification was done on transverse Achilles tendon sections from mice TAM treated at 3 months, injured at 4 months, and stage of analysis post injury is indicated (representative examples shown in Supplementary Fig. 5; n > 3 mice per group per timepoint were analyzed with three 200 × 200 μm square areas quantified per section, >4 sections per mouse; one-way ANOVA, F = 436; ****p < 0.0001; n.s. not significant). J Dot plot showing the average and percent expression of tendon and pericyte markers in our scRNA-seq at 10 dpi. IRC-1 and IRC-2 show similarities in tendon gene expression (such as TnC, Col12a1, and Egr1) with the MBT cluster and the other tendon clusters compared with other cell types. Notably, few genes are shared between IRC-1, IRC-2, and the pericyte cluster.
To determine which cluster contained Axin2+ cells, we examined our single cell RNA-seq datasets for Axin2 and TdTomato transcripts. Axin2+ and TdTomato+ cells were predominantly found in the Scx+ MBT cluster during homeostasis in concordance with our histological and 2-photon microscopy observations (Fig. 5A–D). Axin2+ cells were also found in the Ost and Ent clusters (Figs. 3I and 5B, C), which is consistent with a previous lineage tracing study of enthesis injury44. Although MTJ- and Ent appear similar to the MBT, these distinct cell clusters should be located at the myotendinous and osteotendinous (enthesis) junctions, respectively, and not near the injury site, which is in the middle of the tendon body (for reference please see Supplementary Fig. 2D, E).
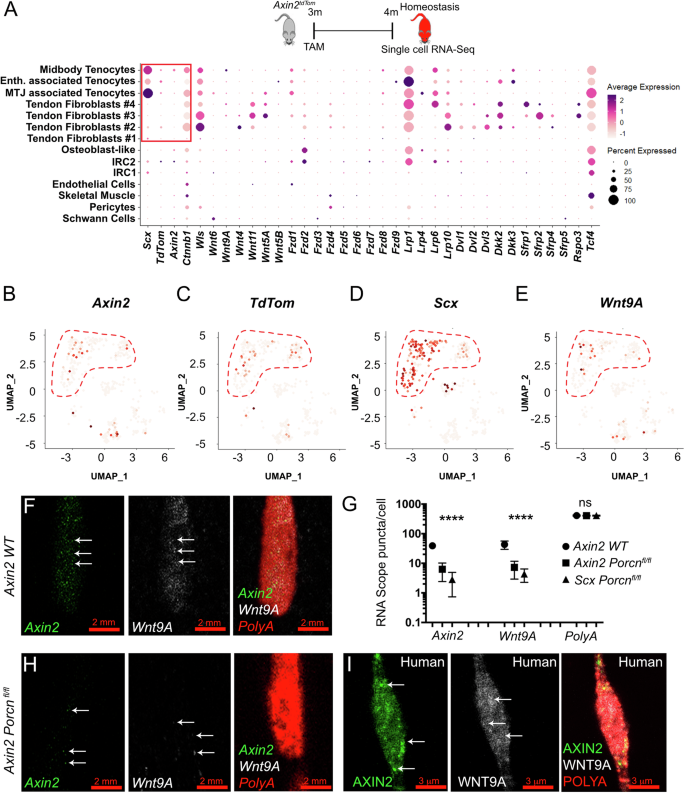
A Cartoon showing the experimental set-up used for the analysis with TAM treatment at 3 months and scRNA-seq of total Achilles tendon cells in homeostasis at 4 months. Single cell RNA-seq dot plot of canonical Wnt pathway components expressed in the different cell clusters during homeostasis; Scx, Tnmd, and TdTom expression is shown. B–E UMAP feature plots of Axin2, TdTom, Scx, and Wnt9A show that these genes are expressed in the MBT cluster during homeostasis (MBT, MTJ, and Ent clusters are outlined in red). F, H smFISH images of one cell containing Axin2 (green), Wnt9a (white), and PolyA (red) puncta in 4-month-old Achilles tendons from wild type (WT) (F) and Axin2:CreERT2; Porcnfl/fl mutants (H; note scale bars are 2 μm). G Quantification of Axin2, Wnt9a, and PolyA RNA puncta per cell in Achilles tendons shows expression of Axin2 and Wnt9A in WT and a decrease in RNA puncta in Axin2:CreERT2; Porcnfl/fl and in Scx:CreERT2; Porcnfl/fl mutants at 4 months of age (n > 8 Achilles tendon per group with n > 20 tendon cells analyzed per tendon; groups consisted of 4-month-old WT, and Axin2TdTom; Porcnfl/fl, ScxTdTom; Porcnfl/fl with TAM given at 3 months; multiple T-test with Holm–Sidak correction was used for statistical analysis; n.s. not significant; ****p < 0.0001). I Representative cell co-expressing AXIN2 (green) and WNT9A (white) in human semitendinosus (hamstring) tendon (I). PolyA (red) was used as an internal expression control (F–I).
Because of the high proportion of Scx+, Axin2+, and TdTomato+ cells in the MBT, we focused our analysis on the MBT cluster. Pseudotemporal reconstruction analysis of the UMAP representation using Slingshot45, inferred a trajectory from the MBT to IRC-1 and IRC-2 clusters (Fig. 4D, E), suggesting that MBT could give rise to cells in these injury-responsive states. IRC-1 and IRC-2 expressed genes known to be upregulated after tendon injury and associated with myofibroblast and mesenchymal identities, including Acta2 and Sox97 (Fig. 4F, G). We focused on the expression of Acta2, which encodes αSMA and is expressed in myofibroblasts, as well as in growing and healing tendons7,46, and Sox9, a gene expressed in skeletal progenitors and chondrocytes47. These genes are expressed in IRC-1 and IRC-2 cells at 10 dpi, respectively (Fig. 4F, G). To determine if Axin2TdTom cells can acquire expression of genes found in IRC-1 and IRC-2 during healing, we used genetic lineage tracing. Immunofluorescent staining of injured Achilles tendons showed a majority of Axin2TdTom cells co-expressing Sox9 and αSMA at 10 dpi, 20 dpi, and 30 dpi whereas uninjured tendons expressed limited amounts of Sox9 and αSMA (Fig. 4H, I and Supplementary Fig. 5A–H). As Acta2 is highly associated with myofibroblasts, which in some studies originate from pericytes or vascular smooth muscle cells48, and because we found Axin2TdTom in vascular-associated cells outside the tendon, we examined the expression of additional genes to determine if IRC-1 and IRC-2 could represent a pericyte-like cell responding to injury rather than a tendon-derived cell. Analysis of gene expression in the different cell clusters after tendon injury shows that IRC-1 and IRC-2 share similarities in gene expression with the tendon clusters, and the MBT in particular (Egr1, TnC, and Col12a1) but do not express several markers that are enriched in pericytes, including Notch3 and Myh11 (Fig. 4J). These results are consistent with Axin2-lineage tendon cells transitioning to IRC-1 and IRC-2 states in healing and support a model in which Axin2+ cells are a major responding cell population in tendon injury. While our pseudotemporal reconstruction analysis was able to infer a trajectory between the MBT and the IRC clusters, it is also possible that other cell populations such as the Axin2+/Scx+ cells from peritendinous regions contribute to the IRC states.
Axin2
TdTom tendon cells are a unique tendon cell population expressing stem/progenitor cell markers
To molecularly examine the Axin2TdTom cells before and after injury, we performed bulk RNA-seq analysis and compared Axin2TdTom to non-Axin2TdTom tendon cells during homeostasis and at 10 dpi. As the total RNA quantity in tendon cells is limiting and to avoid pooling samples or multiple tendon types which individually have a limited number of Axin2TdTom cells, we chose to amplify cohorts of pooled 4-month-old Achilles tendon cells using SMART-seq2 and perform differential expression analysis (Supplementary Fig. 5L, M)49. During homeostasis, Axin2TdTom cells have significantly higher expression of several genes including CD201 (ProCR), Fam102a, Robo2, LepR, and Egfl6 (Supplementary Fig. 5N). CD201 marks stem/progenitor cell populations and is a target of Wnt signaling50. Robo2, LepR, and Egfl6 are expressed in different stem cell populations in the intestine, bone, and epidermis51,52,53, whereas Fam102a has been identified as a late iPSC signature gene54,55 Immunostaining and flow cytometry analysis confirmed enriched expression of CD201 in Axin2TdTom cells (Supplementary Fig. 6B). After injury, Axin2TdTom cells were significantly enriched for Sox9 and Acta2, consistent with our single cell RNA-sequencing and lineage tracing results, and for several tendon genes including Col1a2, Tnmd, and Mkx (Supplementary Fig. 5N). Of note, the previously described tendon sheath stem cell marker Tppp3 was enriched in non-Axin2TdTom cells at homeostasis (lfc = −1.906, p-value = 1.49e-9) (Supplementary Fig. 5N, O), indicating Axin2TdTom cells are distinct from Tppp3+ cells, which have been shown to contribute to patellar tendon healing9. Examination of our single cell data set revealed Tppp3 was expressed in several tendon cell clusters and did not appear restricted to a specific cluster (Supplementary Fig. 5K). This result is consistent with another tendon single cell RNA-seq analysis13. Further expression analysis by smFISH revealed Tppp3-expressing cells throughout the main tendon body and in peritendinous regions (Supplementary Fig. 5P–R) but these cells did not significantly express Axin2, confirming our RNA-seq results. Therefore, Axin2+ cells represent a unique population of injury-responsive cells in the adult tendon that does not overlap with other previously described populations.
Autocrine Wnt regulation of Axin2
TdTom cell state
To interrogate how Axin2TdTom cells are regulated, we examined our RNA-seq datasets and as expected, we found expression of several canonical Wnt signaling components in the Axin2+ cell-containing MBT cluster (Fzd1, Wnt9a, Dkk3, Ctnnnb1, Wls) (Fig. 5A) and enrichment for the Wnt pathway in Axin2TdTom cells (Supplementary Fig. 6A). As Axin2 is a direct target and negative regulator of canonical Wnt signaling56, these data are consistent with the notion that Axin2+ cells are regulated via canonical Wnt signaling. Intriguingly, we identified Wnt9a, a Wnt ligand that promotes the formation of synovial connective tissue cells57, as enriched in the MBT cluster during homeostasis in our single-cell data (Fig. 5A, E) and in Axin2TdTom cells in our bulk RNA-seq data (Supplementary Fig. 5N). Using smFISH, we confirmed that Wnt9a is expressed in Axin2+ cells (Fig. 5F). Based on the co-expression of Wnt9a and Axin2, we sought to test if Wnt signaling from the Axin2TdTom cells themselves is required for their identity in homeostasis. As the MBT cluster is enriched for Scx+ cells, we first tested if Wnt signals originating from a Scx+ tendon cell population are necessary for Axin2+ cell identity. We deleted Porcupine (Porcn), a gene required for Wnt secretion, in Scx-expressing cells using ScxCreERT2-TdTom; Porcnfl/fl mice by giving TAM at 3 months and examining RNA transcripts per cell by smFISH in sectioned tendons at 4 months. We found decreased transcript puncta per cell for Axin2 and Wnt9a (Fig. 5G), suggesting that Wnt ligands originating from Scx+ tendon cells are required to maintain Axin2 expression during homeostasis. To specifically test if Wnt secretion from Axin2TdTom cells is necessary to maintain their identity, we TAM-treated Axin2CreERT2-TdTom; Porcnfl/fl mice at 3 months and examined their tendons at 4 months. We found decreased expression of both Axin2 and Wnt9a upon Porcn deletion in Axin2-expressing cells (Fig. 5G, H). We confirmed that the cells were viable in Porcn mutants as we detected a strong PolyA RNA signal in each cell analyzed, which also standardized our staining assay (Fig. 5F–H). In addition, we found a decrease by flow cytometry in the percentage of Axin2TdTom tendon cells co-expressing ProCR/CD201 from Axin2CreERT2-TdTom; Porcnfl/fl mice compared to wild-type cells (Supplementary Fig. 6B–D). Despite the appearance of similar numbers of Axin2TdTom cells in our tendon sections of Porcn conditional knockout mice, we detected a decrease in total Axin2TdTom cells in mutant compared with control tendons by flow cytometry (Supplementary Fig. 6E, F), suggesting the loss of Wnt signaling affected Axin2TdTom cell survival. Notably, we found that some populations of non-Axin2-expressing cells expressed other Wnt ligands during homeostasis (e.g,. TF2–4; Fig. 5A), suggesting that Wnt secretion from Axin2TdTom and ScxTdTom cells is uniquely necessary for maintaining Axin2 and Wnt9a expression. Together, these data indicate that active Wnt secretion from Axin2TdTom tendon cells is required to maintain their cell state and Wnt9a expression, suggesting a positive feedback loop maintains their own identity.
Next, we sought to determine if this unique Axin2 + /Wnt9a+ cell is present in human tendons. We obtained healthy human hamstring tendons from patients ranging in age from 18 to 22 years and examined AXIN2 and WNT9A expression. Within the main tendon body, we observed a subset of cells expressing AXIN2 and WNT9A (11.7% ± 3.48% and 12.6% ± 4.7% cells per total nuclei per 200 µm2 area, respectively; n = 4), and a majority of the AXIN2+ cells (86.6% ± 7.6) co-expressed WNT9A (Fig. 5I). Together, these findings suggest that a similar AXIN2+ cell population exists in human tendons.
Loss of Wnt signaling from Axin2
TdTom cells impairs healing
As loss of Porcn either in Scx– or Axin2-expressing cells negatively affects the ability of the Axin2 cells to maintain their identity, we next tested if this loss affects their ability to respond to tendon injury. First, we deleted Porcn from the Scx-expressing cells (ScxCreERT2-TdTom; ; Porcnfl/fl) by giving TAM at 3 months and performing partial excisional tendon injuries at 4 months. Examination of ScxCreERT2-TdTom; Porcnfl/fl mice revealed a severely impaired healing response with virtually no Scx-lineage cells, reduced Scx-GFP expression, and a lack of matrix re-organization in the injured area at 30 dpi compared with wild-type injured tendons (Fig. 6A, B, and E; Supplementary Fig. 8A, and Supplementary Movie 10). During the healing process in wild-type tendons, we observed increased gene expression of Axin2, Scx, Col1a2, and Ki67 by RT-qPCR relative to uninjured contralateral control Achilles tendons (Supplementary Fig. 7). Genes associated with a mature tendon matrix, such as Fibromodulin (Fmod) and Connexin 43 (Cx43)58 were significantly upregulated at later healing time points, 20 dpi and 30 dpi, likely signifying cell differentiation and matrix maturation events (Supplementary Fig. 7A). Unlike the control injured tendons, ScxCreERT2-TdTom; Porcnfl/fl injured Achilles tendons did not display dynamic changes in the expression of tendon transcription factors (Scx, Mkx), matrix molecules (Col1a2, Cola3a1), proteins found in mature tendons (Fibromodulin, Connexin 43), proliferation (MKi67), and genes associated with mesenchymal progenitors or differentiation towards other cell fates (Sox9, Runx2) at 10 dpi, 20 dpi, and 30 dpi (Supplementary Fig. 7B), suggesting a severely blunted healing response.
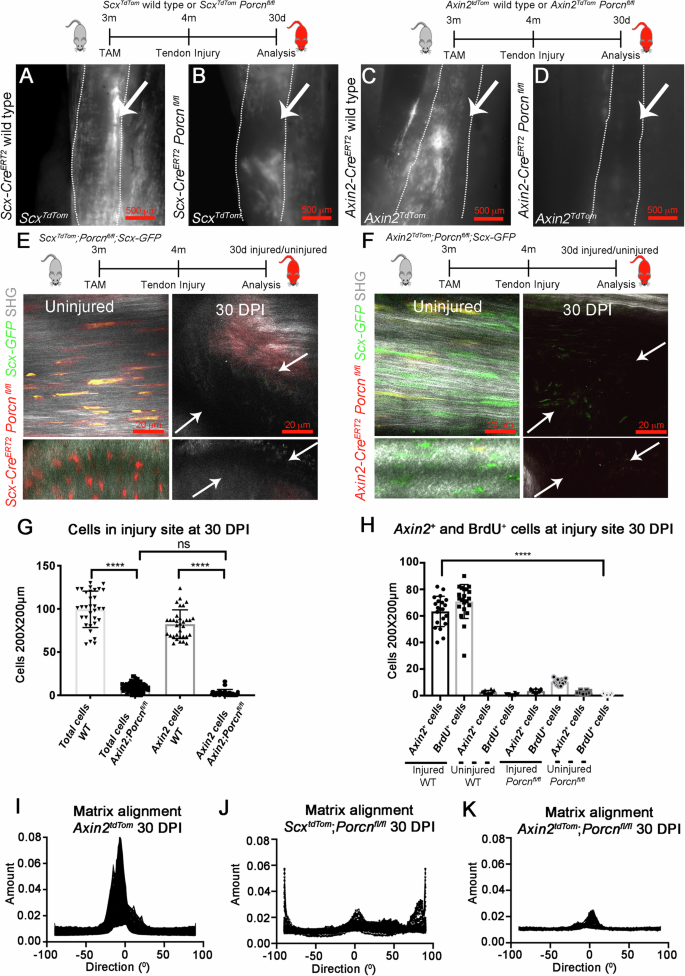
Cartoon representations of the experimental design and mouse genotypes with TAM treatment at 3 months and tendon injury at 4 months are shown. A–D Whole-mount fluorescent images of the whole Achilles tendon at 30 dpi (outlined by dotted line; muscle attachment is towards the top and bone attachment at bottom) ScxTdTom and Axin2TdTom cells localize to the injury site (arrows) in WT controls (A, C), but not in Scx:CreERT2TdTom; Porcnfl/fl or Axin2:CreERT2TdTom; Porcnfl/fl mutants (B, D). Two-photon fluorescent and SHG images of contralateral uninjured Achilles tendons (E, left) and injured 30 dpi Scx:CreERT2-TdTom; Porcnfl/fl ; Scx-GFP mutant Achilles tendons (E, right), and of uninjured (F, left) and injured 30 dpi Axin2-CreERT2; TdTom; Porcnfl/fl; Scx-GFP mutant tendons (F, right). A–F (n = 6 mice per group per time point). G Quantification of total cells and Axin2TdTom (Axin2 cells) cells per 200 × 200 μm square area in the injury site in WT and in Axin2-CreERT2-TdTom; Porcnfl/fl mutant Achilles tendons at 30 dpi (n = 4 mice per group, with three 200 × 200 μm square areas quantified per section and n > 3 sections analyzed per mouse per time point; Unpaired T-test with Welch correction was used; n.s., not significant; ****p < 0.0001). H Quantification of Axin2TdTom and BrdU+ cells per 200 × 200 µm square area in injured and uninjured Achilles tendons from Axin2TdTom WT compared with Axin2:CreERT2-TdTom; Porcnfl/fl mutants at 30 dpi (n = 5 mice per group with quantifications from three 200 × 200 µm square areas per section, and three sections per tendon; unpaired T-test with Welch correction was used; ****p < 0.0001). I–K Samples were analyzed for directionality using Fourier spectrum analysis and the FIJI directionality plugin was used to compute the histograms indicating the amount of structures and its direction in each image. The X-axis shows the direction of the structure in degrees, and the Y shows the sum of directions from each slide of the 2P images. I Axin2TdTom mice, there was more alignment into one coherent direction, whereas in J ScxCreERT2-TdTom; Porcnfl/fl and in F Axin2CreERT2-TdTom; Porcnfl/fl mice, the directionality was mostly un-orientated (n > 3).
To test if Wnt secretion specifically from the Axin2-expressing cells is required for tendon healing, we analyzed Axin2CreERT2-TdTom; Porcnfl/fl Achilles tendons at several stages after injury using the same TAM treatment and injury strategy. Similar to ScxCreERT2-TdTom; Porcnfl/fl injured tendons, the healing response was significantly impaired with disruption to the SHG signal, reduced cell proliferation, and abnormal morphology in healing Axin2CreERT2-TdTom; Porcnfl/fl Achilles tendons compared to controls at 30 dpi (Fig. 6C, D, and F–H, Supplementary Fig. 8A, and Supplementary Movie 11). Within the injury site, we observed a virtual loss of all Axin2TdTom cells, as well as significantly fewer total cells and cells incorporating BrdU in Axin2CreERT2-TdTom; Porcnfl/fl tendons compared to injured controls (Fig. 6G, H). Further examination revealed an increase in CD45+ cells at the healing site of Axin2CreERT2-TdTom; Porcnfl/fl tendons compared to wild type (WT) tendons at 30 dpi (Supplementary Fig. 8D, E,), suggesting that some of the cells present in Porcn mutants are blood cells. In addition, there was no significant increase in Scx, Mkx Col1a2, MKi67, Fmod, and Sox9 expression along with several other genes in the injured compared to contralateral uninjured Achilles tendons of Axin2CreERT2-TdTom; Porcnfl/fl mice (Supplementary Fig. 7C). Similarly, we observed significantly reduced numbers of Axin2TdTom cells that expressed Sox9 and αSMA in injured Axin2CreERT2-TdTom; Porcnfl/fl Achilles tendons compared to controls (Supplementary Fig. 8B, C), suggesting the IRC-1 and IRC-2 cell types are not present in Porcn mutants. In addition, Fourier transform methods were used to quantify matrix organization by assessing angles of deviation from the main direction of alignment59,60. We found disorganized matrix orientation upon loss of Porcn by Axin2CreERT2 or ScxCreERT2 compared with controls (Fig. 6I–K and Supplementary Fig. 9). The similarities in the severity of the healing phenotype observed in the Scx- and Axin2-mediated Porcn conditional loss-of-function supports a model in which Axin2TdTom cells are the major Wnt-secreting subset of the Scx cells and that they regulate their injury response in an autocrine manner. As the Axin2TdTom cell identity is affected in Porcn mutants, we cannot rule out that their inability to mount a healing response results from their loss of competence to respond to injury. Moreover, the lack of other Axin2(Neg) cell populations responding to injury in Axin2CreERT2-TdTom; Porcnfl/fl indicates the Axin2+/Scx+ cells through their secretion of Wnt signals, are central to the recruitment of virtually all tendon cells in healing.
Discussion
Our work identifies a unique Axin2+/Scx+ tendon cell population that mobilizes upon injury to promote healing and, through its secretion of Wnt, serves as the central organizer of the healing response. Our single cell and genetic analysis reveals heterogeneity in tendon cell populations and positions Axin2+/Scx+ tendon cells as key responders to tendon injury via regulation by their own Wnt signals (Fig. 7). Axin2+ cells have multilineage differentiation potential, are enriched for previously described markers of stem and progenitor cells including ProcR/CD201, and expand extensively in vitro, while at the same time maintaining higher expression of markers of tendon fate. During homeostasis in vivo, Axin2+ cells are latent, dividing less frequently than their non-Axin2TdTom counterparts, yet are poised to respond to injury, a characteristic of several stem and progenitor cell populations61. Consistent with this concept, Axin2TdTom cells are the major cell population responding to tendon injury. They infiltrate the injury site, proliferate, and differentiate into elongated Scx-GFP tenocytes. Upon transplantation into injured immunodeficient mice, significantly expanded Axin2TdTom cells differentiate into tenocytes and self-renew into Axin2+ cells in the host tendons. Therefore, we propose that Axin2TdTom cells are the endogenous cell source of the previously described TDSPCs14.
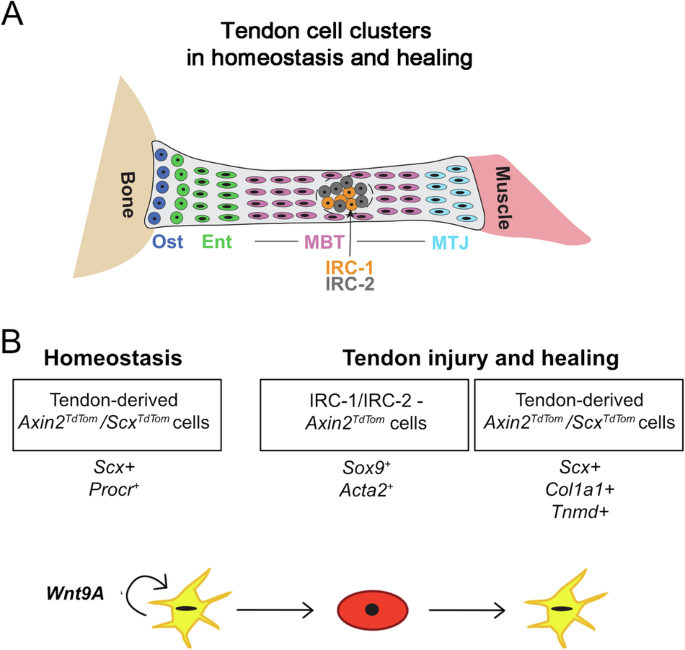
Cartoon depiction of the identified cell types in the tendon during homeostasis and injury with a focus on relevant clusters (A). In our model, B Axin2+/Scx+ tendon cells require their own Wnt signal to maintain their Procr/CD201+ and Axin2+ state in homeostasis. Upon injury, our single-cell RNA-seq and genetic lineage tracing show that Axin2+ cells adopt an injury-responsive state (IRC-1 and IRC-2), expressing Sox9 and Acta2. They robustly migrate, proliferate, adopt a rounded morphology, and then differentiate into tenocytes expressing common markers of tendon fate (Scx, Col1a1, and Tnmd). These processes are entirely dependent upon the secretion of their own Wnt ligands, likely Wnt9a.
Transcriptomics, single molecule in situ hybridization, and functional deletion of Porcn in Axin2TdTom and ScxTdTom cells revealed that Axin2+/Scx+ cells rely on their own secreted Wnt ligands to maintain their identity and injury response. The importance of Wnt secretion from the Axin2TdTom cells to maintain Axin2+ cell state in homeostasis is underscored by their loss not only of ProcR expression but also of both Axin2 and Wnt9a upon Axin2CreERT2– or ScxCreERT2-mediated deletion of Porcn. This result suggests that the secretion of Wnt by Axin2+/Scx+ cells is simultaneously maintaining Axin2 identity and the expression of its own Wnt ligand, thereby providing a positive feedback loop to ensure the maintenance of the cell type. Notably, Wnt signaling is important for proliferation and maintenance of tendon fates in vitro as the addition of Porcupine inhibitors resulted in decreased proliferation and Scx expression and an increased propensity to form cartilage. Autocrine regulation is a mechanism employed by other stem and progenitor cell populations3,62. As Wnt ligands act as short-range signals63, the Axin2+/Scx+ cell provides its own niche-like factors that act as a self-sustaining feedback loop. Autocrine signaling is uniquely suited for sparsely situated tendon progenitor cells that have to organize healing in the dense ECM environment. The Axin2+/Scx+ cell population presents an intriguing paradigm for how matrix-rich tissues regulate their maintenance and repair. In periods of homeostasis, Axin2+ tendon cells are quiescent and require their own secreted Wnt signals to maintain their identity and upon injury, initiate the healing response. This system is similar in some ways to the muscle in which Pax7+ satellite cells are a distinct quiescent population that activates upon injury64. We propose that the function of Axin2+/Scx+ cells is to maintain tendon fate in homeostasis through their fidelity to tendon gene expression programs yet retain the ability to activate and orchestrate repair upon injury.
Our work also positions Axin2+/Scx+ cells as key early initiators of tendon healing through their secretion of Wnt. We detect Wnt9a expression in Axin2+ MBT cells during homeostasis (Fig. 5), and upon injury, our bulk RNA-seq shows significant enrichment for Wnt9a compared with Axin2(Neg) cells (Supplementary Fig. 5). We also observe increased Axin2 transcript levels in injured relative to uninjured contralateral tendons, suggesting there is active Wnt signaling during injury (Supplementary Fig. 7A). It is interesting to note that reports in the liver reveal an alternative mode of regulation after injury in which there are no specialized subsets of progenitor or stem cells. Instead, all hepatocytes upregulate Axin2 and proliferate upon injury65,66,67. In contrast to this scenario, our results favor a model in which the Axin2TdTom cells are a pre-defined subset of the Scx+ population. Adult Axin2TdTom cells are distinct from Axin2(Neg) cells in their proliferation history since birth (Fig. 2D), suggesting they are uniquely regulated. In addition, specific deletion of Porcn in the Axin2TdTom or ScxTdTom cells one month prior to the injury dramatically affects the cellular response to injury. Porcn-deleted Axin2TdTom and ScxTdTom cells fail to respond to the injury but are readily observed in the contralateral uninjured tendon. At the injury site, there are fewer Axin2(Neg) cells and an increase in CD45+ cells, indicating that loss of Porcn in the Axin2TdTom cells more broadly affects the healing response. Axin2+ and Scx+ cells represent a subset of the total cells in the tendon, approximately 10%, of the cells in the fascicles (Axin2TdTom) or 8% of the cells in peritendinous regions (ScxTdTom) (Fig. 2C and Supplementary Fig. 1 and 2). Notably, our conditional loss of function results indicate that the remaining Axin2(Neg) or Scx(Neg) tendon cells cannot compensate for the loss of Wnt signals from the Axin2TdTom cells or ScxTdTom cells, strongly suggesting that the overlapping Axin2+/Scx+ double positive cells are the key Wnt secreting cells in the tendon. The lack of healing response in Porcn conditional mutants also demonstrates that cells cannot acquire Axin2+/Scx+ cell identity from other Wnt sources or that Wnt secretion from other sources cannot compensate for loss from Axin2TdTom or ScxTdTom cells. The Porcn-deleted Axin2TdTom and ScxTdTom tendons also display a deficient healing response with disorganized matrix deposition at 30 dpi (Fig. 6I–K and Supplementary Fig. 9). Although this suggests a significantly disrupted healing response, it remains unclear if there are functional impairments to the healed tissue at later stages. Mechanical testing of these tendons is an important area of our future study. Together, our Porcn loss of function data underscore the importance of the Axin2+/Scx+ cells as essential for their Wnt secretion and organization of the early healing response.
How the adult Axin2+ cells acquire their identity is currently unclear. Recent studies in a neonatal tendon regeneration model have shown that Axin2-lineage cells contribute to tendon healing26. Our transcriptomic and genomic analysis of the first postnatal month68 combined with our lineage tracing at P2 (Fig. 2A–C and Supplementary Fig. 3), show increased Wnt pathway components and increased Axin2+ cells, respectively, at early neonatal stages. The decline in canonical Wnt signaling occurs concomitantly with a decrease in tendon cell proliferation69; marked matrix expansion70, and a loss of regenerative healing46,71. It is possible that the adult Axin2+ cells are descendants of a subset of these earlier Axin2+ cells that remain latent yet retain the potential for regenerative healing in the adult. We have used the H2B-GFP label-retaining assay to determine that the P60-labeled Axin2+ cells descended from a population that divided less frequently since birth than the Axin2neg cells at that stage. This is based on the higher percentage of H2B-GFP+ cells and higher intensity GFP label in the Axin2+ cells that were labeled at P60. Recent work has indicated that there may be caveats with the H2B-GFP label retaining assays as degradation of the H2B-GFP protein was shown to occur over week-long time-scales in the hematopoietic system72,73. While this could suggest that protein degradation is a possible mechanism of decreased H2B-GFP signal, our assays in this and previous works17 show stable H2B-GFP expression in tendon cells over month-long postnatal chase periods. This is consistent with other reports of H2B-GFP labeling lasting over 6 months in post-mitotic retinal cells74, suggesting that H2B-GFP degradation and stability may be different in different cell types. Additionally, our previous work also used a complementary method of EdU and BrdU labeling to confirm our results from the H2B-GFP system at similar stages in the mouse17. Therefore, we believe that the H2B-GFP label-retaining assays show reduced proliferation of the Axin2+ cells compared with Axin2neg cells at postnatal stages with decreasing numbers of Axin2+ cells from postnatal to adult stages. Whether Axin2+ cell number remains constant at later stages is currently unknown, but it would be interesting to investigate if changes in Axin2+ cell number or activity underlies age-related increases in tendon injury and disease.
Previous studies have shown that stem and progenitor cells from the tendon sheath are involved in tendon healing7,8,9. We provide evidence for a distinct cell population, marked by Axin2 and Scx, that are quiescent during homeostasis, yet display progenitor cell behavior and are capable of activating upon injury to directly orchestrate the healing response. As Scx is mainly expressed in tenocytes in the main tendon body and a majority of the cells expressing Axin2, Scx, and Wnt9a are found in the MBT cluster, it is possible that the main responding cell population is an Axin2+/Scx+ cell in the main tendon body. However, given that Axin2TdTom cells and ScxTdTom cells are detectable in peritendinous regions, we cannot exclude the possibility of Axin2+/Scx+ peritendinous cell involvement in tendon healing, which would be consistent with previous reports of sheath-residing progenitor cells7,9. The lack of sheath/paratenon-specific markers complicates their definitive identification in the context of our Axin2CreERT2-TdTom lineage tracing strategy. Similar to another Achilles tendon single-cell sequencing study13, we found that expression of Tppp3, a previously described sheath marker, was not restricted to a specific tendon cluster and was expressed in both peritendinous regions and the main body of the Achilles tendon (Supplementary Fig. 5K, N, and O–R), making it difficult to resolve which cluster may represent the sheath-like cell population. As the prior work also described these cells as Tppp3+ and Pdfgra+9, we examined the clusters and found co-expression of Tppp3 and Pdgfra in a subset of clusters, Ent, TF-2, TF-3, and TF-4 (Supplementary Fig. 5J, K). It is possible that one of these clusters, particularly TF-4, which significantly expresses both genes, represents the previously described sheath/paratenon population. Nevertheless, our expression analyses show that Axin2TdTom cells are distinct from Tppp3-expressing cells and our Porcn loss-of-function data positions Axin2TdTom cells as providing an upstream signal necessary for cell recruitment and proliferation at the injury site. Therefore, our work shows that the Axin2+/Scx+ cells are distinct from previously described Tppp3-expressing cells and are key responders to tendon injury through their secretion of Wnt signals. These conclusions are based on our bulk and single cell RNA-seq data and the similarities in lineage tracing and in phenotypes resulting from Porcn deletion using ScxCreERT2-TdTom and Axin2CreERT2-TdTom. These results are also consistent with previous studies that have shown contribution from Scx-lineage and Scx-GFP+ cells to tendon injury18,34.
In this work, we developed a transplantation assay using an alginate system to assess the ability of Axin2TdTom cells to incorporate and contribute to healing in tendons of immunodeficient host mice as opposed to transplantation to non-tendon locations that were previously used14. For these experiments, it was necessary to significantly expand Axin2TdTom tendon cells for generating the alginate gel-Axin2TdTom cell construct. Following transplantation into injured tendons, we observed differentiated Tnmd+ Axin2TdTom cells and Axin2TdTom cells that expressed Axin2+, providing strong evidence of Axin2TdTom cell self-renewal. A drawback of our approach was the need to pool large numbers of cells, which made clonal expansion unfeasible. Measurements of clonality and long-term repopulating potential are the gold standard for stem cell assays to functionally establish that a single cell is capable of both differentiation and self-renewal. As a hypocellular and dense matrix tissue, many approaches used in other systems can be challenging to apply to the tendon. However, we believe our transplantation assay is an important first step towards performing similar tests of distinct tendon cell populations in the tendon proper and can serve as the foundation for future functional assays.
The identification of similar AXIN2+/WNT9A+ cells in human tendons opens new potential therapeutic avenues to treat tendon disease and injury. Notably, the percentage of AXIN2+ cells in humans is similar to adult mouse tendons, representing approximately 10% of the cell population. As we used human hamstring tendons for this analysis, it suggests Axin2+ cells may be broadly present in other tendon types. It is particularly interesting that Axin2+ cells are enriched for Wnt9a in humans and mice as prior work has shown that this and other Wnt ligands have important roles in both promoting synovial connective tissue formation and suppressing chondrogenic potential in the context of in joint formation57. Notably, specific loss of Wnt9a and Wnt4 in limb mesenchyme results in ectopic mineralization in adult tendons and their attachments75, likely due to it so function in cartilage and bone formation76,77. As the appearance of ectopic bone and cartilage is associated with adult tendon overuse and injury46, it is interesting to speculate that perhaps aberrant regulation of Wnt signaling by the Axin2+ cells could underlie these pathologies. Not only do Axin2+ cells represent a new potential source for tissue engineering strategies, but they are also an exciting novel therapeutic target for tendon injury and disease.
Methods
Mouse husbandry
All mouse husbandry and experiments were performed according to American Veterinary Medical Association (AVMA) guidelines and were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee (IACUC: 2013N0000062). The following mouse strains were used: Axin2tm1(cre/ERT2)Rnu/J (Jax Cat# 018867);27,28 H2B-GFP (Col1a1:tetO-H2B-GFP;ROSA-rtTA35; Scx-Cre78, Scx-GFP32,79 Scx-CreERT2); Porcupinefl/fl (129S-Porcn tm1.1Vdv/J, Jax Cat# 020994); Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (Jax Cat# 007909); Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (Jax Cat# 007676), and Foxn1Nu/Nu (JAX 000819).
Injuries
Excisional Achilles tendon injuries were performed using a 0.3 mm biopsy punch as described69,80. The incisions were sutured with 6-Ethilon nylon sutures, and tendons were harvested at specific times after injury. Mice were given anesthetics and analgesics for the injury studies and were euthanized according to the AVMA guidelines. All work was approved by the MGH IACUC (2013N0000062).
Administration of TAM
Two miligram of TAM from stock of 20 mg/ml in corn oil (Sigma Aldrich Cat# T5648) was given intraperitonially with each mouse receiving this 3 times every other day beginning on the day described in the text (P2, P60, or 3 months of age; approximately 80 mg/kg body weight). After the administration of Tam, we waited at least a month for the Tam to be washed out to avoid any persistent activation of CreERt2.
BrdU administration and staining
BrdU was injected at a concentration of 150 mg/kg (Sigma Cat#B5002) as described17 from the day of the injury until the collection of the tendons. For BrdU immunostaining, sections or cultured cells underwent antigen retrieval and immunostaining using anti-BrdU (1:100; Abcam Cat# 6326). For cell culture, BrdU was added with a final concentration of 10 μM BrdU. For BrdU immunostaining, antigen retrieval with a primary anti-BrdU Antibody (1:100; Abcam Cat# 6326) and a secondary antibody (Abcam Cat #150167) were used.
H2B-GFP induction
H2B-GFP expression was induced by DOX administration (2 mg/ml in drinking water with sucrose and 150 μM one-time injection into timed pregnant females at E10 and drinking water was continued until birth). Upon birth, DOX is removed for the chase period (starting at P0), and the GFP is diluted in proportion to cell division. Axin2TdTom and Axin2(Neg) cells were isolated from the hindlimbs and forelimbs at indicated stages and analyzed as described in the flow cytometry section.
RNA extraction and RT-qPCR
RNA extraction and RT-qPCR of mouse Achilles tendons were done according to17,69. Fold change was calculated using the dCt method81 and statistics were performed on the dCt values.
RT-qPCR primer sequences are as follows:
|
Transcript |
Forward sequence (5’–3’) |
Reverse sequence (5’–3’) |
|---|---|---|
|
Gapdh |
TGTTCCTACCCCCAATGTGT |
GGTCCTCAGTGTAGCCCAAG |
|
Col1a2 |
CCAGCGAAGAACTCATACAGC |
GGACACCCCTTCTACGTTGT |
|
Col2a1 |
CAGGGCTCCAATGATGTAGA |
TCTTCTGTGATCGGTACTCG |
|
Acan |
TTGCCAGGGGGAGTTGTATTC |
GACAGTTCTCACGCCAGGTTTG |
|
Tnmd |
AACTCCACCTCAGCAGTAGTCC |
TTTCTTGGATACCTCGGGCCAGAA |
|
Fmod |
CATGGCAACCAGATTACC |
AGATATAAGGCCGTGAGG |
|
CNX43 |
TTGACTTCAGCCTCCAAGG |
AATGAACAGCACCGACAGC |
|
Scx |
AAGTTGAGCAAAGACCGTGAC |
AGTGGCATCCACCTTCACTA |
|
Ki67 |
AGCAAGCCAACAGAATTTCCAG |
TATCTTGACCTTCCCCATCAGG |
|
Sox9 |
AGTACCCGCATCTGCACAAC |
TACTTGTAATCGGGGTGGTCT |
|
Mkx |
AGTGGCTTTACAAGCACCGT |
TTTGACACCTGCACTAGCGT |
|
Col3a1 |
TGACTGTCCCACGTAAGCAC |
GAGGGCCATAGCTGAACTGA |
|
Axin2 |
GACCGACGATTCCATGTCCA |
GCGTGGGTTCTCGGAAAAT |
Postnatal RNA-seq
Extraction of intact total RNA from whole tendons was performed17,69. Between 7 and 9 biological replicates (P0-8 mice, P7-7mice, P14-7 mice, P21-7 mice, P28-8 mice, and P35-9 mice) per weekly time point (P0-P35) passed quality measures, yielding a total of 46 samples spanning 6-time points. RNA-seq library preparation was completed in a single batch on an Apollo 324 NGS library prep system (IntegenX) and the PrepX mRNA library protocol (Takara Bio) in the Harvard Bauer Core Facility. Single-end 75 bp reads were sequenced on an Illumina NextSeq 500 using a high-output 75-cycle kit (Illumina) in the Harvard Bauer Core Facility. Sequenced reads were pre-processed and quantified using Salmon82. DESeq2 was used to determine differential expression68. All downstream analyses were performed in R (R Core Team 2019).
Postnatal ATAC-seq
Scx-lineage cells were isolated from Scx-Cre; TdTom+ mice via enzymatic digestion and FACS sorted into replicates of 5000 cells each. Transposition and library preparation were performed using a modified version of the Fast-ATAC protocol. Paired-end 42 bp reads were sequenced on an Illumina NextSeq 500 using a High-Output 75-cycle kit (Illumina) in the Harvard Bauer Core Facility. Peaks were identified from Bowtie2 aligned reads using MACS283 and all downstream analyses were performed in R68 (R Core Team 2019) using the following packages: DiffBind84, ChIPseeker85, clusterR, ComplexHeatmap86, and ClusterProfiler87.
Axin2
TdTom SMART seq
Freshly isolated tendons were collected from 4-month-old mouse forelimbs and hindlimbs or from injured Achilles tendons (N > 10 mice; Tam at 3 months; collected at 4 months in homeostasis, or injured at 4 months and collected at 10 dpi) and dissociated as described17,69. The cells were sorted using BD FACSARIA II. CD31-APC, and CD45-APC Cy7 cells were excluded (BD Cat# 551262, Cat# 557659). Axin2TdTom and Axin2(Neg) cells were sorted separately into 96-well plates (Eppendorf, 951020401) into 50 μl of TCL buffer (QIAGEN 1031576, supplemented with 1% β-mercaptoethanol), 200–1000 cells of Axin2TdTom or Axin2(Neg) cells were sorted into each well, and stored at −80 °C. Libraries from the cell lysates were generated using the SMART-Seq protocol88, with some modifications in the reverse transcription step89. The 96 well plates with the cell lysates were thawed on ice, spun down at 1500 RPM for 30 s, and mixed with Agenocourt RNAclean XP SPRI beads (Beckman Coulter) for RNA purification. Purified RNA was resuspended in 4 μl of Mix-1, desaturated at 72 °C for 3 min, and placed immediately on ice for 1 min before 7 μl of Mix-2 was added. Reverse transcription was carried out at 50 °C for 90 min, followed by 5 min incubation at 85 °C. Fourteen microliter of Mix-3 were added in each well and the whole transcriptome amplification step was performed at 98 °C for 3 min, followed by 21 cycles at (98 °C for 15 s, 67 °C for 20 s, and 72 °C for 6 min), and the final extension at 72 °C for 5 min. cDNA was purified with Agencourt AMPureXP SPRI beads (Beckman Coulter) as described89. Quality control steps were performed on samples before library construction as follows: concentration measurements using Qubit dsDNA high-sensitivity assay kit; and cDNA size distribution using the High-Sensitivity DNA Bioanalyzer kit. Libraries were generated using the Nextera XT library prep kit (Illumina). Combined libraries were sequenced on NextSeq 500 sequencer (Illumina), using paired-end 38 base reads, and resulted in approximately 30 million 75 bp reads per sample. Data from all samples were processed using an RNA-seq pipeline implemented in the bcbio-nextgen project (bcbio-nextgen,1.1.6a-b’2684d25’). Gene expression was quantified using Salmon (version 0.13.1) using the 0 mm transcriptome (Ensembl 92, 2018-10-10). To identify differentially expressed genes in a pairwise manner, the statistical package DESeq2 (version 1.24.0) was utilized. Genes were classified as differentially expressed based on a 2-fold change in expression value and false discovery rates below 0.05.
Single-cell RNA-seq
Achilles tendons were freshly isolated from four adult mice (4 months of age) with 8 tendons total for homeostasis, and from three adult mice (4 months of age) with three tendons total for 10 days post-injury. Tendon cells were dissociated as described17,69 and sorted using BD FACSARIA II. CD31-APC, CD45-APCCy7 and TER-119/Erythroid APC-Cy7 cells were excluded (BD Cat#551262, Cat# 557659, Cat#560509). Immediately following FACS, cells were loaded to the 10× platform using Chromium next GEM single cells 3’ reagents kit V3.1. Libraries were prepared using the single-cell 3’ Reagent kit (Version 3.1 10× Genomics, Pleasanton, CA, USA). Following library preparation and quantification, libraries were sequenced by the MGH Core using either the NextSeq 2000 or HiSeq 2500 to generate 200 million reads per sample. A total of three samples were analyzed: total cells at homeostasis, total cells at 10 dpi, and sorted tdTomato+ cells at 10 dpi from Axin2-creERT2; flox-stop-flox-tdTomato mice (Tam given at 3 months). Raw fastq files for each sample were first trimmed using Trimmomatic90 to remove low-quality reads. Trimmed reads were subsequently processed using Cell Ranger and the output was analyzed using Seurat v4.091. First, to broadly identify which cell types were captured, all three samples were individually filtered to remove cells with <100 genes, <200 UMIs, low complexity (log(genesperUMI) < 0.8), and a high mitochondrial ratio (>0.2). The samples were then normalized using the SCTransform function and clustered to identify and remove CD45+ cells that should have been removed by sorting. All three samples were then integrated together (using top 3000 variable features and top 30 principal components). PCA and UMAP dimensionality reduction was performed followed by clustering at various resolutions (a resolution of 1.0 was deemed appropriate). Major cell types present were classified into tendon, Schwann cells, skeletal muscle, pericytes, or endothelial cells based on previously characterized cell type markers.
To classify tendon cells at higher resolution, tendon cells were a subset (based on Col1a1 expression) from each individual sample and then integration was performed (using the top 3000 variable features and top 30 principal components). PCA and UMAP dimensionality reduction was performed along with subsequent clustering at different resolutions (a resolution of 1.0 was ultimately determined suitable). Different tendon cell populations were classified based on the expression of known markers for cells in different regions of the tendon during homeostasis (0 dpi) (ex. Scx for all tenocytes, Col22a1 for MTJ-associated tenocytes, Pthlh and Sox9 for Ent). Four tendon clusters that could not be readily identified based on marker expression were deemed as tendon fibroblast cells (TF1–4). UMAP representations, dot plots, and heatmaps were generated via Seurat or SCANpy92. Pseudotime trajectory analysis was performed on a subset of the integrated Seurat object (total homeostasis and 10 dpi samples only) using Slingshot45 with the root of the trajectory set to the MBT cluster. To examine the expression of Wnt pathway components in the tendon with respect to all of the other cell types, cells from the tendon clusters identified here were manually assigned onto the UMAP from the Seurat object containing all cell types via their barcode. From this re-mapping, 61 cells across all three samples that were originally assigned to “tendon” in the larger Seurat object containing all cell types remained unassigned indicating these cells had not been included in the higher resolution tendon cell analysis (i.e., 18 cells for the homeostasis sample, 6 cells for the 10 dpi sample, and 38 cells for the Axin2-tdtomato sorted sample). A re-examination of where these cells clustered in each individual sample revealed these cells were not included in the original tendon cell analysis because they did not consistently cluster with tendon-like cells. Because these cells could not be confidently re-assigned to a corresponding tendon cluster during the re-mapping step and to keep the analysis consistent, these 61 cells were removed only for this specific analysis. Expression of Wnt pathway component expression was then examined on the subset of cells from homeostasis.
Flow cytometry
Tendon cells were isolated from distal forelimb and hindlimb tendon tissue of mice at the age indicated in the text and figure legends for Figs. 2 and 3A–L17. Achilles tendons were isolated for the multilineage experiments in Fig. 2M and Supplementary Fig. 3C, in the single cell RNA-seq in Figs. 3–5, and in the transplantation assay in Fig. 4H. We enriched Axin2TdTom tendon cells by excluding CD31+ and CD45+ cells prior to analysis (BD Cat# 551262, Cat# 557659). DAPI or Calcein Blue AM staining was used to exclude dead cells or select for live cells. For each experiment, the gates were set using negative and positive controls for each fluorescent color used. To evaluate GFP intensity in the H2B-GFP flow cytometry experiments, we used fluorescent reference beads to calibrate samples between experiments (molecular probes Cat# C16508) as described17. The following antibodies were used for flow cytometry analysis: CD90.2, CD44, Sca-1 (BD Cat# 56257, Cat# 563970, Cat# 561021), and ProCR (CD201) (Biolegend Cat# 141505).
smFISH and immunohistochemistry
We treated 8 µM paraffin sections of injured and uninjured Achilles tendons with TEG buffer for 5–6 h for antigen retrieval, used proteases 3, four treatment steps (RNA-Scope, ACD Cat#322340), and continued according to the protocol (RNAScope) for detection of the mRNA with the following probes: Hs-Axin2 (Cat# 400241); Hs-Wnt9A (Cat# 457931); Mm-Wnt9A (Cat# 405081); Mm-Axin2 (Cat# 400331); Mm, Hs-PolyA (Cat# 318631). Immunohistochemistry was performed on cryo-sections as described17 with anti-mCherry (Origene/Sicgen AB0040-200), anti-Sox9 (Millipore Cat# AB5535), and anti-αSMA (Sigma Cat# C6198). For characterization of Axin2 and Scx-lineage populations during homeostasis in Supplementary Fig. 1, 6-month-old uninjured Axin2TdTom or ScxTdTom mice were dosed with TAM at 1-week prior to tissue collection. The limbs were collected and processed for 2-photon imaging as described below (Supplementary Movies 1–5) or fixed overnight at 4°, brought up a sucrose gradient, and embedded in OCT for cryosectioning (n = 3 for each mouse line). Limb tissue was sectioned at 10 µM using cryotape (Kawamoto method) and immunofluorescence staining was performed using an anti-tdTomato antibody (Origene Cat# AB0040). Stained sections were then imaged using confocal microscopy (Leica SP8 X (HC PL APO 40X/1.3W CORR, HCX APO 63X/0.9W U-V-I CS2). For each mouse line, 3–5 regions of interest along the tendon were quantified on 1–2 stained cryosections per limb. Fascicular, peritendinous, and surrounding non-tendinous tissue was defined based on anatomy (see Supplementary Fig. 1 for examples).
Cell culture
We isolated tendon cells from 4-month-old mice after Tam at 3 months. On day 0, Axin2TdTom and Axin2− cells were either plated together in cell culture dishes prior to analysis of surface marker expression by flow cytometry (day 10) or were plated separately for RT-qPCR analysis (day 10). DMEM (Gibco Cat# 119 65-092) with 1% P/S (Corning Cat# 30-002-cl), Fetal Bovine Serum (FBS, Gibco Cat# 97068), and 1% Hepes (Gibco Cat# 15630) was replaced every 2 days. To inhibit Porcupine and Wnt secretion, we added Wnt 974 dissolved in DMSO to a final concentration of 2.5 μM (Sigma Cat# 1243244-14-15) or DMSO alone to Axin2TdTom cells. Also, on day 0 of plating, BrdU was added to both conditions every other day for 5 days total. The cells were fixed and immunostained for BrdU and TdTomato. For RT-qPCR the cells were collected and processed with RNA Zymo Kit according to69.
Human samples
Human tendon samples were collected from patients aged 18–22 undergoing ACL reconstruction using an autograft from the semitendinosus (hamstring) tendon We analyzed >20 sections per sample, with n = 4 separate human tendon samples. As we only used discarded human tissue and properly de-identified all samples, informed consent was waived. Research involving human research material has been performed per the Declaration of Helsinki under IRB# 2013P001931 with the approval of the MGB Institutional Review Board.
Imaging
Two-photon microscopy was used to image control and injured tendons as described17. To standardize the laser power “Bright Z” was adjusted. The images were taken with optical slices of 0.4 µm with a 25× wet lens (XPlan N 25× WMP) on an Olympus 2P microscope FLOWVIEW FVMPE-RS. Confocal microscopy was used to image immunohistochemistry and smFISH of Achilles tendon sections using a Leica SP8 × (HC PL APO 40×/1.3 W CORR, HCX APO 63×/0.9W U-V-I CS2). Zeiss AxioZoom V16 was used for whole-mount hindlimb and Achilles tendon images and Zen software was used for processing. For Cell culture images, we used Leica DM IL inverted microscope, and pictures were taken in 10× and 20× magnification.
Multilineage differentiation assays
Axin2TdTom cells were isolated at 4 months (Tam at 3 months) from Achilles tendons and cells from each mouse (n = 5) were cultured and expanded separately for 60 days. After obtaining sufficient Axin2TdTom cell numbers, we performed adipogenic, osteogenic, and chondrogenic assays according to93.
For Adipogenesis assays, after seeding in a 6-well plate, Axin2TdTom cells were fed with a growth medium consisting of 10% FBS, 1% Pen/Strep, and 1% HEPES in DMEM/F12. Cells were expanded to near confluence in a 37 °C humidified incubator with 5% CO2. The growth medium was replaced with a pre-warmed Adipogenesis Differentiation Medium. This was done according to the protocol in the StemPro Adipogenesis Kit (Gibco MAN0000693). Cells were fed every 2–3 days. Once the cells adopted an adipogenic-like phenotype (14–21 days), we removed the differentiation medium, rinsed twice with DPBS, and fixed the cells with 4% PFA for 30 min. PFA was removed and cells were rinsed twice with DPBS and 1 ml of 0.2% w/v Oil Red O Solution was added after filtering through a 0.22 uM filter membrane. Oil Red O preparation: we prepared a working solution that is two parts 0.5% stock solution (0.5% w/v solution in 100% isopropanol) and three parts distilled water. We discarded the Oil Red O and rinsed twice with 60% isopropanol and once with ddH2O. We added 1 ml Hematoxylin to the well, incubated at RT for 2 min, and rinsed with ddH2O until clear.
For Osteogenesis assays, after seeding in a 6-well plate, Axin2TdTom cells were fed with a growth medium consisting of 10% FBS, 1% Pen/Strep, and 1% HEPES in DMEM/F12. Cells were expanded to near confluence in a 37 °C humidified incubator with 5% CO2. The growth medium was replaced with a pre-warmed Osteogenesis Differentiation Medium. This was done according to the protocol in the StemPro Osteogenesis Kit (Gibco MAN0000694). Cells were fed every 2–3 days. After 21 days, we removed the differentiation medium, rinsed twice with DPBS, and fixed the cells with 4% PFA for 30 min. We removed the PFA, rinsed twice with ddH2O, and added 1 ml of 2% Alazarin Red S solution in water for 5 min. We discarded the Alizarin Red S and rinsed it with ddH20 until clear. We added 1 ml Hematoxylin to the well, incubated at RT for 2 min, and rinsed the wells with ddH2O until clear.
For Chondrogenesis assays 5 μl micromasses with 100 cells per μl were created in 6 well plates. Axin2TdTom cells were fed a growth medium consisting of 10% FBS, 1% Pen/Strep, and 1% HEPES in DMEM/F12 and incubated for 2 h in a 37 °C humidified incubator with 5% CO2. The growth medium was replaced with a pre-warmed Chondrogenesis Differentiation Medium, according to the protocol in the StemPro Chondrogenesis Kit (Gibco MAN0000696). Cells were fed every 2–3 days. After at least 14 days, we removed the differentiation medium, rinsed twice with DPBS, and fixed the cells with 4% PFA for 30 min. PFA was removed, the cells were rinsed twice with ddH2O, and 1 ml of 1% Alcian Blue Solution (1% w/v solution in 0.1 N HCl) was added for 30 min. After discarding Alcian Blue solution, we washed the cells three times with 0.1 N HCl, and once with ddH2O. We added 1 ml of Hematoxylin to the well, incubated at RT for 2 min, and rinsed with ddH2O until clear.
Transplantation assay
We isolated 100–500 Axin2TdTom cells from Achilles tendons from 4-month-old mice (Tam at 3 months) by FACS. We excluded dead cells and CD31 + /CD45 + /TER119+ cells. Axin2TdTom cells from each mouse (two Achilles tendons, n = 5) were plated and expanded separately for 60 days. Prior to transplantation, we performed flow cytometry on Axin2TdTom cells to confirm they were TdTomato+ and quantify cell number. After obtaining approximately 4 million Axin2TdTom cells, we combined Axin2TdTom cells with alginate-based hydrogels and transplanted them into injured tendons of host immunodeficient mice Foxn1Nu/Nu (JAX 000819). Under general anesthesia with isoflurane, both hind limbs were shaved, prepped with 1:1 betadine/alcohol solution, and draped. Buprenorphine and bupivacaine (around the intended surgical site) were administered 30–60 min prior to the procedure. A 5 mm longitudinal incision was created in the hindlimb only in the skin from the anatomical location of the Achilles myotendinous junction to the osteotendinous junction, thus providing exposure to the plantaris longus and Achilles’ tendons. Microscissors were used to separate the plantaris longus from the Achilles tendon prior to performing a full-thickness partial width (50%) injury to the central mid-substance of the Achilles tendon with a 0.3 mm biopsy punch. Briefly, a 1% oxidized RGD modified (DS20) alginate gel (VLVG, NovaMatrix) was synthesized94,95. Ultrapure alginate samples were first dissolved in Dulbecco’s Modified Eagle Medium at 2.857% (w/v). Seven hundred microlitre of this alginate solution was syringe mixed with a second syringe containing 100 µl of cell suspension (40 million cells/ml) and 200 µl of calcium sulfate dihydrate (240 mM stock) The mixture was then quickly cast between glass plates separated at a 1 mm thickness and left to crosslink for 20 min. Gels were then punched into 3 mm diameter disks and rinsed in DMEM prior to implantation on the same day. The skin was closed using 4-0 vicryl and the animal returned to a regular cage activity. After 10 days and 20 days, The Achilles’ tendons were harvested for evaluation.
Quantification and statistical analysis
The statistical methods, p values, and numbers analyzed are indicated in the corresponding Figure legends. In brief, for comparisons between flow cytometry data either a two-tailed unpaired T-test with Welch correction was used (Figs. 2C and 3G–K) or one-way ANOVA (Fig. 2D). For qRT-PCR analysis, Multiple T-test with the two-stage step-up method of Benjamini, Krieger and Yekutieli was used. For analysis of quantification of cell counts, a T-test with Welch correction was used. For quantifications of cell culture (Fig. 3), cells isolated from an individual mouse were considered one biological replicate, and cells were counted in defined square areas as indicated in the figures with at least five random areas measured per biological replicate indicated. For quantification of cells in tendon sections, cells were counted in defined square areas as indicated with at least three areas measured per section and 3–5 sections per tendon with n > 4 mice per experimental group. For statistical analysis of smFISH experiments (Fig. 5F, G), multiple T-tests with Holm–Sidak correction were used.
Responses