A multiplex microarray lateral flow immunoassay device for simultaneous determination of five mycotoxins in rice

Introduction
Rice is an important staple crop for more than half of the world’s population1. Like other cereals, rice grains are highly susceptible to fungal contamination, particularly during the ripening stage (pre-harvest) and storage (post-harvest)2. Aspergillus, Fusarium, and Penicillium are the three main fungal genera that have been reported to frequently infect rice2,3,4. Under favorable temperature and humidity conditions, these fungal species produce toxic metabolites known as mycotoxins5. The most prevalent mycotoxins in rice and rice-based products worldwide are aflatoxin B1 (AFB1), T-2 toxin (T2), zearalenone (ZEA), deoxynivalenol (DON), fumonisin B1 (FB1), ochratoxin A (OTA), citrinin, patulin, and sterigmatocystin2,3,5.
These toxic compounds can induce a wide range of adverse health effects in humans and animals6. For instance, the trichothecenes (DON and T2) and ZEA have been reported to interfere with protein synthesis and reproductive processes, respectively7,8. AFB1 can induce teratogenic effects in humans and animals; thus, they have been classified under Group 1 Carcinogens by the International Agency for Research on Cancer (IARC)9. Chronic exposure to FB1 has been demonstrated to induce esophageal cancer in humans10. IARC has also classified FB1 as possibly carcinogenic to humans (Group 2B)9,10. To ensure the safety of foods and feeds in the supply chains, several countries have set maximum permitted levels (MPLs) for mycotoxin residues in agricultural commodities11. In the European Union, the established MPL for unprocessed rice is 5 µg/kg for AFB1, 10 µg/kg for the sum of AFs, 100 µg/kg for the sum of T2 and HT-2 toxin, 100 µg/kg for ZEA, 1250 µg/kg for DON, and 5 µg/kg for OTA11.
To detect and quantify these mycotoxins, gas and liquid chromatography coupled to tandem mass spectrometry (GC–MS/MS and LC–MS/MS) and other detectors such as diode array and flame ionization are commonly used in various agricultural commodities12,13. Furthermore, these analytical instruments are used as a confirmatory method and are considered to be the gold standard for regulatory purposes due to their sensitivity, selectivity, robustness, and accuracy13. They do, however, require specialized skills, complicated sample, preparation procedures, expensive laboratory solvents and consumables, and are highly susceptible to matrix interferences14,15,16. Furthermore, they are generally time-consuming and labor-intensive, making them unsuitable for rapid analysis and on-site monitoring of mycotoxin contamination14,15,16.
Over the past decades, lateral flow immunoassays (LFIAs) have emerged as the most used point-of-care testing methods, primarily due to their simplicity, affordability, rapidity, and portability15,17,18,19,20. Nevertheless, most commercially available LFIAs for mycotoxin analysis can only detect a single mycotoxin at a time, with complex sample preparation steps. Previously, we have developed a microarray lateral flow immunoassay (µLFIA) for detecting five mycotoxins. However, its application was limited due to the lack of an easy sample preparation step with the capacity to extract multiple mycotoxins simultaneously and an affordable portable microarray reader21. Up to now, sample preconcentration or purification is usually required before sample application to the available LFIAs; however, these steps are generally expensive and laborious with the use of organic solvents (methanol, acetonitrile, hexane, and chloroform) that are toxic, flammable, and harmful to humans and the environment17,18,19. In addition, the quantitation of mycotoxin levels detected by LFIAs usually requires the use of a costly laboratory benchtop device or bulky and complicated reader needing specialized/trained individuals due to the multiple stages and time-consuming procedures to generate mycotoxin concentrations dataset. Furthermore, most LFIA readers only store the generated data internally or require connection to a local network; thus, are not suitable for on-site testing where portability and wireless connection are very vital.
This study, therefore, developed and validated a low-cost innovative portable and easy-to-use electronic reader together with a multiplex microarray-based LFIA device and an environmentally positive (green) sample extraction procedure for the simultaneous detection and accurate quantitation of trace levels of five important regulated mycotoxins (AFB1, T2, ZEA, DON, and FB1) commonly detected in rice worldwide22,23,24. The portable reader is small, inexpensive, and sensitive, equipped with its own power supply for on-site/in-field accurate quantitative measurement of mycotoxins in agricultural products. The sample extraction process is also fast and suitable for multi-mycotoxin extraction using a green solution containing a mixture of polyethylene glycol (PEG) and ethanol.
The entire testing procedure is very simple. Milled rice sample is mixed with a green extraction solvent before the supernatant is diluted and directly dropped into the microarray strip fabricated with mycotoxin standards. The microarray-based immunoassay detects mycotoxins by means of a direct competitive approach utilizing a novel luminescent dye conjugated to goat anti-mouse antibody (GAM) as a fluorescent reporter. The portable microarray reader captures and processes the microarray fluorescence spots to quantify the concentration of each mycotoxin in the contaminated rice. Overall, this new analytical detection technology enables the identification of mycotoxin-contaminated rice products on-site and improves the environmental footprint of mycotoxin testing, resulting in a much more effective mitigation measure and reducing the associated food waste and economic losses associated with product rejections.
Results
Optimization of microarray lateral flow immunoassay multiplex detection conditions
The detection capacity of a lateral flow immunoassay is generally dependent on a wide range of factors, including the type of membrane, buffers, and specificity of antibodies employed, as well as antigen and antibody concentrations25,26. In this study, the following factors were considered and optimized to improve the mycotoxin detection capacity of the μLFIA: concentration of antibodies, concentration of GAM-M424 conjugate, and microarray running buffer composition.
The monoclonal antibodies used for the development of this assay were produced in-house and have been previously demonstrated to possess high specificity in terms of binding respective mycotoxins27. The optimal concentration of each monoclonal antibody for simultaneous determination of AFB1, T2, ZEA, DON, and FB1, up to 40 combinations of different concentrations (ranging from 3 to 100 µg/mL) of antibodies were investigated using the μLFIA strip test with AFB1-BSA, T2-BSA, ZEA-BSA, DON-BSA, and FB1-BSA spots. As depicted in Fig. S1, the highest spot signal was obtained using 15 µg/mL of AFB1 antibody, 21 µg/mL of T2 antibody, 12 µg/mL of ZEA antibody, 4.5 µg/mL of DON antibody, and 18 µg/mL of FB1 antibody. Different optimal concentrations of these mycotoxin-specific antibodies can be attributed to different levels of affinities of the selected antibodies28.
Different concentrations of GAM-M424 (25, 75, 100, 110, 120, 130, 140, and 150 µg/mL) on microarray spot intensity and background noise were tested, and 100 µg/mL of GAM-M424 was selected as the optimal concentration resulting in the highest spot intensity with low background noise (Fig. S2).
The effect of different concentrations (0.1%, 0.2%, 0.5%, and 1%) of Tween 20, BSA, and Triton X-100, along with percentages and MW of PEG, on µLFIA strip signal intensity and background noise were evaluated. The PEG types tested included MW100 (5%, 10%, and 20%), MW600 (5%, 10%, and 20%), MW3350 (5%, 7.5%, and 10%), MW6000 (0.5%, 1%, 2.5%, and 5%), MW8000 (1%, 2.5%, 3%, 4%, 5%, and 6%), and MW20,000 (1%, 1.5%, 2%, 2.5%, and 5%). The optimal results were obtained using 5% MW6000, 4% MW8000, and 2% PEG MW20,000 (PEG 20K) (Fig. S3).
Based on mycotoxin extraction recovery results and the observed µLFIA strip spot intensity, a running buffer with a mixture of 3.9% PEG 20K, 2% tween 20, and 1% BSA was finally selected as it provided the highest spot intensity and flow rate, with low background noise after incubation of only 15 min at room temperature.
Green mycotoxin extraction procedure
A wide range of natural or non-toxic and biodegradable solutions, including ionic liquids, deep eutectic solvent, and various non-ionic surfactant solutions (Tween 20, Triton X-100, and PEG), were examined during the preliminary study to identify a substance or mixture of compounds that are compatible with the μLFIA strip components and can also be used for the extraction of target analytes. Following a series of mycotoxin spiking experiments using LC–MS/MS, PEG 20K was found to meet the pre-defined criteria (i.e. non-toxic, biodegradable, and compatible with mycotoxins and μLFIA strip). As illustrated in Fig. 1, 0.1% of PEG 20K supplemented with 20% ethanol in 10 mM acetate buffer (pH 5.0) gave the best recovery for all the five target mycotoxin compounds. Ethanol (20%) was added to the PEG solution because it improved the flow rate of the μLFIA strip and yielded high recovery for ZEA and AFB1, which are generally known to be insoluble in aqueous solution when compared to T2, DON, and FB1.
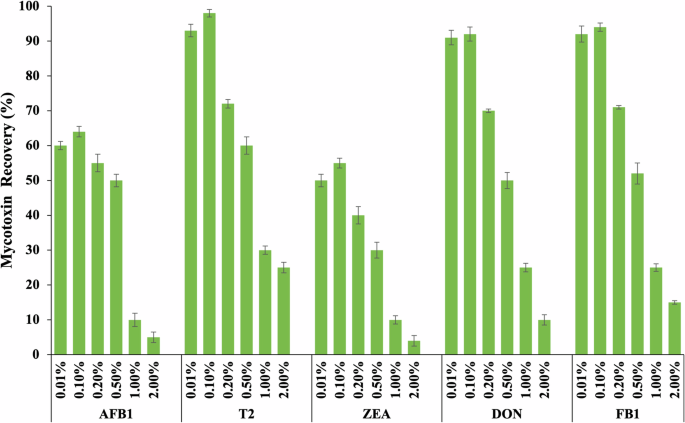
Percentage recovery of AFB1, T2, ZEA, DON, and FB1 from different concentrations of polyethylene glycol (PEG 20K) solution spiked with multiple mycotoxins. The PEG solution was spiked at 20 ng/mL for each toxin and analyzed using LC–MS/MS. Error bar represents a standard deviation value (n = 3). The letters represent a significant difference at p < 0.05 in each treatment of mycotoxin.
The recovery of mycotoxins at various time intervals was also investigated using the developed green solution (Fig. 2). As there were no significant differences (p > 0.05) with regard to mycotoxin recovery at 0.5, 1, 2.5, 5, and 10 min. However, to ensure that all mycotoxins were extracted from the rice sample, a 1 min period was selected for the simultaneous extraction of mycotoxin in the unprocessed rice sample. While a sample preparation method based on PEG-sodium citrate aqueous two-phase system has been previously reported for the extraction of AFB1, DON, and OTA in feed29, to the best of our knowledge, this is the first study to describe the use of an ultra-fast (1 min) and PEG-based extraction procedure for the determination of five mycotoxins, including AFB1, T2, ZEA, DON, and FB1, in rice samples via the use of a μLFIA.
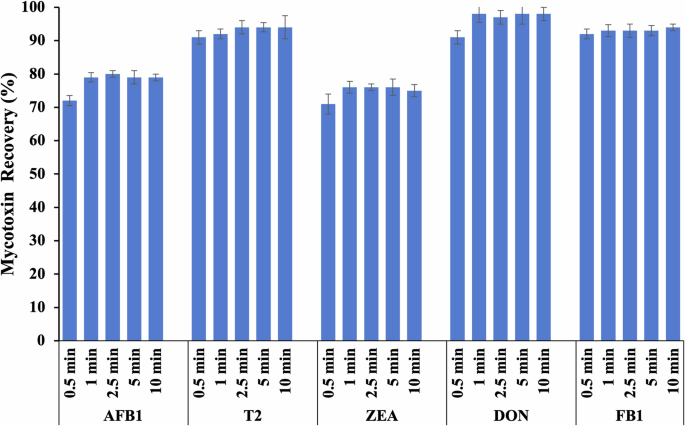
Percentage recovery of AFB1, T2, ZEA, DON, and FB1 from rice samples at different time points using an extraction solution containing 0.1% PEG 20K with 20% ethanol. The rice samples were spiked at 20 ng/g and analyzed using LC–MS/MS method. Error bar represents a standard deviation value (n = 3). The letters represent a significant difference at p < 0.05 in each treatment of mycotoxin.
Microarray portable reader for the lateral flow immunoassay strip test
A cost-effective and portable microarray reader was developed and validated for rapid on-site mycotoxin detection. The reader provides a user-friendly operation with a few simple steps. Additionally, the reader was constructed entirely using off-the-shelf components, thereby significantly reducing the reader’s cost (estimated at US$ 1100). The component list is shown in the Supplementary Fig. S4. Its dimensions and weight are 15.0 × 18.5 × 17.5 cm and 2.0 kg, respectively. These features position the developed reader as a bridge between laboratory-based mycotoxin detection and on-site applications, enhancing accessibility and usefulness for a broader group of users and ultimately contributing to safer food. With the optimized software and hardware, we have achieved the readout of microarray signals of different mycotoxins in just 1 min while maintaining good signal repeatability and robustness.
Performance of the microarray-based lateral flow immunoassay detection platform
The optimized assay conditions were used to evaluate the specificity of the developed µLFIA strips for singleplex and multiplex detection. We tested both target and non-target compounds through a competitive assay strategy. Each µLFIA strip consisted of five mycotoxin conjugates (AFB1-BSA, T2-BSA, ZEA-BSA, DON-BSA, and FB1-BSA), positive control (RAG antibody) and negative control (BSA) as shown in Fig. 3. We tested the capacity of the strip to accurately detect single mycotoxins using 11 compounds (AFB1, AFB2, AFG1, AFG2, AFM1, T2, HT-2, ZEA, DON, FB1, and OTA), while the multiplex detection accuracy was tested with different combinations of multiple mycotoxins (2-plex, 3-plex, 4-plex, and 5-plex). The sample signals were compared to the signals obtained from blank samples (i.e., without any mycotoxin addition).
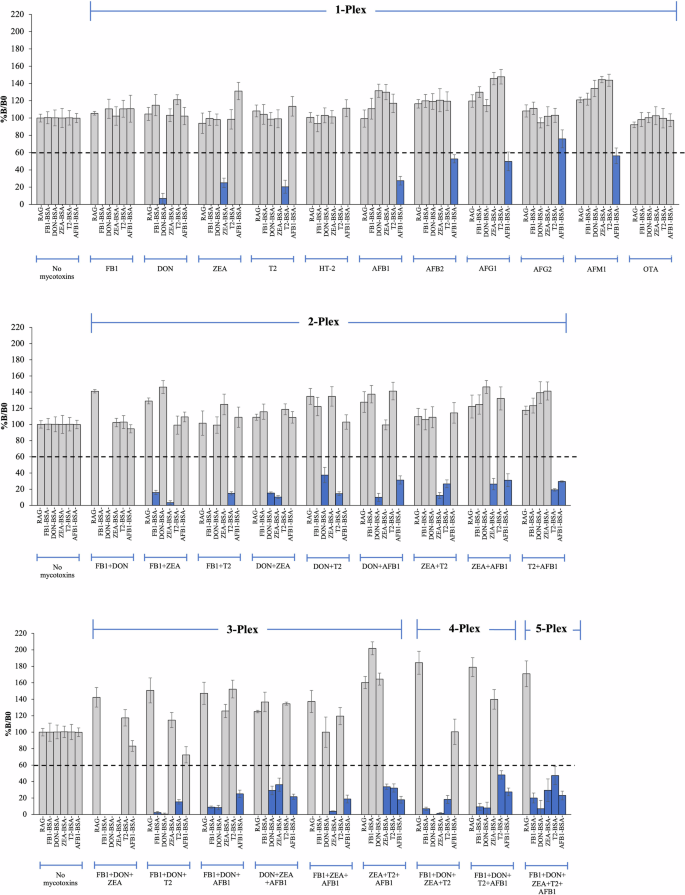
Individual mycotoxins, namely FB1, DON, ZEA, T2, HT-2, AFB1, AFB2, AFG1, AFG2, AFM1, and OTA, were tested for single mycotoxin detection (1-Plex). Each data was plotted as an average spot intensity of 12 replicate spots, with error bars indicating standard deviation. The blue bars represent a positive result after testing with spiked mycotoxins. Dotted line represents a cut-off value, which is an average spot intensity of 12 replicate spots from a mycotoxin-free sample strip minus 3 times standard derivation values.
As shown in Fig. 3, the µLFIA strip test showed specific detection for each of the five target mycotoxins (AFB1, T2, ZEA, DON, and FB1). However, a cross-reactivity was observed for AFB1, AFB2, AFG1, AFG2, and AFM1, as well as T2 and HT-2. This is not unexpected as these chemical compounds have similar structures or functional groups. This also indicates that the assay has the capacity to detect and estimate total aflatoxins (i.e., the sum of AFB1, AFB2, AFG1, AFG2, and AFM1 and the sum of T2 and HT-2) present in a sample. For multiplex detection, µLFIA showed high specificity and accuracy when tested against different combinations of multiple mycotoxins (Fig. 3). For instance, the intensities of T2-BSA and FB1-BSA spots were reduced in a sample containing a mixture of only T2 and FB1. As illustrated in Fig. 3, similar results were observed for all the target mycotoxins either in the 2-plex, 3-plex, 4-plex, or 5-plex assay. Overall, these results showed that the developed µLFIA was able to accurately detect single and multiple mycotoxins with a high degree of specificity.
After investigating the μLFIA specificity and confirming the absence of cross-reactivity, a matrix-matched calibration curve was constructed using a wide linear range of multi-mycotoxin concentrations to compensate for matrix effects and to determine the sensitivity of the assay. The EU regulatory limits and levels of mycotoxins commonly found in agricultural products in Southeast Asia were used to select the range of concentrations for each of the toxin22,23. The logarithmic, linear regression equations and standard curves for AFB1, T2, ZEA, DON, and FB1 are shown in Table 1 and Fig. 4, respectively. The R-squared or coefficient of determination values obtained for the target compounds were >0.97, showing a very good linearity of the calibration curves.
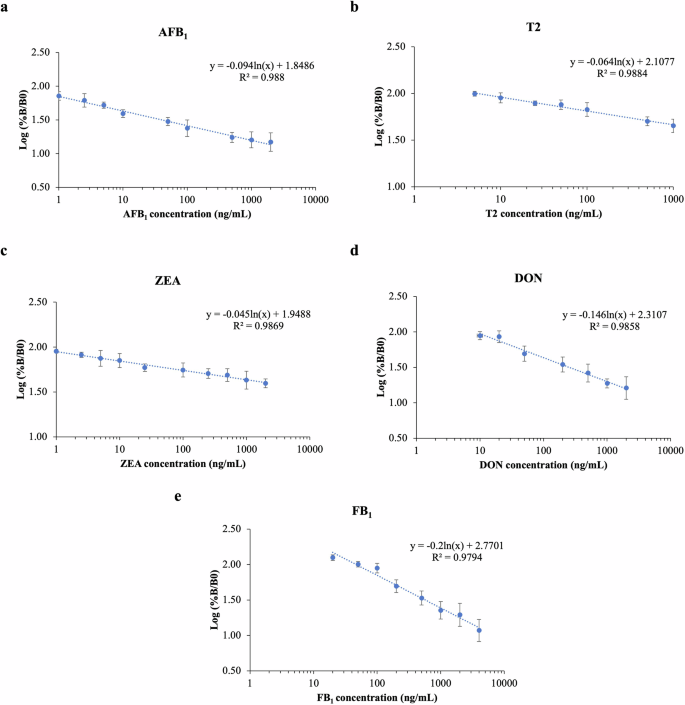
The linear regression and equation for a AFB1, b T2, c ZEA, d DON, and e FB1. Each standard data point was plotted as an average intensity of 12 replicate spots from three strips with an error bar indicating a standard deviation value.
Mycotoxin-free rice samples were spiked at three different concentrations to reach the target concentration of 100, 200, and 500 µg/kg. However, following extraction and dilution, the final concentrations of mycotoxins in the samples were 10, 20, and 50 µg/kg (~10-fold dilution). The precision of µLFIA was evaluated in terms of intra-assay (repeatability) and inter-assay (reproducibility). Table 2 outlines the validation performance of the developed rapid assay in rice samples. The LODs obtained for AFB1, T2, ZEA, DON, and FB1, respectively, were 1.89, 1.21, 1.39, 1.17, and 0.56 µg/kg. The LOQ was 6.3 µg/kg for AFB1, 4.02 µg/kg for T2, 4.62 µg/kg for ZEA, 3.91 µg/kg for DON, and 1.97 µg/kg for FB1 (Table 1). The average recovery values ranged between 77% and 127%; the intra-day and inter-day %RSD values ranged from 5% to 25% for the target mycotoxins. These LODs and LOQ values demonstrated the method’s high capacity, stability, and sensitivity for screening large numbers of food and feed samples, effectively reducing the number of contaminated samples before consumption. Moreover, these values are generally considered satisfactory and in line with the performance criteria laid down by the European Commission Regulations for rapid screening methods30. In addition, the recovery values obtained for target mycotoxins using the μLFIA strip were relatively similar to LC-MS/MS results, confirming the accuracy and reliability of the developed rapid assay (Table 2).
Discussion
Previously, our team has successfully developed a μLFIA for simultaneous detection of five mycotoxins, but the previous work was limited to lab-based experiments where no quick and effective sample preparation protocol was available to simultaneously extract multiple mycotoxins in real samples. In addition, the previous work was devoid of a portable detector hindering its full exploitation21. Therefore, this study strived to address those pitfalls by developing a point-of-care device, consisting of a developed μLFIA strip, a green extraction solution, and an inexpensive portable reader. The developed system was further validated for the simultaneous detection of five mycotoxins (AFB1, T2, ZEA, DON, and FB1) in real food samples, such as rice, to ensure its usefulness in the real world in terms of rapid detection and quantification of co-contaminated samples in a sustainable manner.
To develop μLFIA for five mycotoxins detection, antibody concentrations were optimized. The highest spot signal was selected as the optimal concentration for these mycotoxin-specific antibodies, attributed to varying affinities of the selected antibodies28. The sensitivity of an immunoassay is largely influenced by specific monoclonal antibodies31,32,33.
Labeled immunoassays, including fluorescence, bioluminescence, colloidal gold, organic dye, and magnetic beads, are widely used to detect binding events in many biomedical and diagnostic analyses31. In this study, a recently developed organic fluorescent dye, M42434, was used as a reporter molecule for this assay21. To benchmark with commercial dyes (e.g. fluorescein aldehyde (FAM-aldehyde) and cyanine3 (Cy3)), which are extensively applied in medical diagnostics, imaging, forensics, highlighters, and writing utensils35,36, M424 exhibits higher optical quantum yield than FAM-aldehyde and Cy321,35,36 (Table S1). In this study, 100 µg/mL of M424-GAM was selected as the optimal condition for the developed μLFIA strip due to M424’s significantly larger Stokes shift compared to FAM-aldehyde and Cy3. This presents a significant benefit in that M424 can be easily detected by a simple reader without using a complex and expensive light filter and permits quantitative analysis for any multiplex detecting platform. Moreover, M424 emits at the near red region, while FAM-aldehyde emits green light, presenting an advantage for M424 to be used as an alternative signal reporter in the application that a test sample has an autofluorescence in the green region such as aflatoxins.
For green extraction procedures, organic solvents, including methanol, hexane, acetonitrile, and acetone, are generally used to extract mycotoxins and other chemical contaminants from various agricultural commodities37. These solvents are primarily obtained from non-renewable resources (fossil-based) and pose a substantial risk to human and animal health38. In addition, organic solvents significantly contribute to the generation of hazardous wastes, constituting an important source of environmental hazards, expensive chemical waste disposal for laboratories and volatile organic compound emissions39.
Recognizing the environmental impact of traditional extraction methods, it is crucial to consider the sustainability of sample preparation. The sample preparation metric of sustainability (SPMS) was followed to evaluate the greenness, environmental friendliness, or sustainability of the developed sample preparation technique40. Parameters were grouped into four different categories according to the type of information that they provide: (i) sample information (sample amount, either volume for liquid samples, or weight for solid samples), (ii) extractant information (amount, nature, and reusability), (iii) sample preparation procedure information (number of steps, extraction time, need of additional steps after extraction, and sample throughput), and (iv) energy consumption and waste. The detail of the evaluation for the standard method (AOAC), commercial method, and the present work is demonstrated in Table S2.
It was found that our sample preparation gave a higher score metric of sustainability than that of the sample preparation used by AOAC for analysis of the same mycotoxin, such as aflatoxins. Extracting solvents based on mixed organic solvents, including methanol, which is well-known as a hazardous reagent to humans and the environment, was used in the standard method41,42. When compared to a sample preparation method explained in a commercial kit for mycotoxin detection, the score for the sample preparation developed in the manuscript implied fairly a similar sustainability result to the commercial one. Additionally, for this developed system, a single sample extraction is performed for five mycotoxin analyses simultaneously, while an extraction for commercial detection is done for only one mycotoxin analysis. Due to the different chemical properties of different mycotoxins, analysis of many mycotoxins by commercial kit may require more than one extraction.
To improve the sustainability of mycotoxin testing, a non-toxic, biocompatible, and non-flammable PEG solution presents as a good alternative43,44 for a green extraction solution in line with the green analytical chemistry principles45. Therefore, the multiplex detection of five mycotoxins can dramatically reduce not only analysis time but also energy consumption. For these reasons, the present sample preparation and microarray-based immunoassay technique are considered eco-friendly technology.
PEG is a synthetic, biocompatible, and hydrophilic polyether compound46. Due to its non-toxicity and high-water solubility, PEG is commonly used for drug delivery and as a surfactant, emulsifier, laxative, ointment, and dispersing agent in medical, pharmaceutical, and cosmetic industries46,47. PEG is also authorized as a food additive in many countries, including the EU and USA48. Ethanol is classified as an environmentally preferable solvent as it can be produced from renewable sources, such as sugars, starches, and lignocellulose49.
The performance of our assay was compared to other recently published microarray-based multiplex methods50,51,52,53,54, as shown in Table 3. Our developed platform successfully detected the regulated five mycotoxins simultaneously within 17 min, whereas other microarray platforms took over 60 min. PEG, aimed at reducing environmental toxicity in both sample preparation and analytical steps, was effectively utilized for extracting the five mycotoxins. However, the LODs in this method were slightly higher than in other microarrays, but the LODs remained below MPLs. As demonstrated in Table 3, our analytical device developed and validated in this study is faster and incorporates more integrated components while offering a greener and simpler extraction procedure.
Finally, a multiplex and eco-friendly microarray lateral flow immunoassay device was successfully developed and validated for the simultaneous determination of five mycotoxins in rice. The system utilized a novel organic fluorescent dye M424 as a signal reporter and was equipped with an eco-friendly extraction protocol with an inexpensive portable device. This platform showed excellent performance: simple, rapid, accurate, eco-friendly, and high sensitivity. From the extraction until signal readout, the entire protocol is complete within 17 min. The assay recovery values were acceptable in the range of 77–127%, and the relative standard deviation (%RSD) was between 5% and 25%. The LODs for the five mycotoxins detection were shown to be in the range of 0.56–1.89 μg/kg. Importantly, the major components used for the fabrication of this analytical system, including monoclonal antibodies, fluorescent dye, solvents, and microarray reader, were all produced in-house. This opens the possibility to upscale the production of point-of-care devices that are cost-effective for end-users such as farmers and researchers in academia and the food industries. While this system presents many innovative advantages, future efforts will be dedicated to addressing a few challenges, such as scaling up the production of the system, optimizing protocols for other matrices, and including other important regulated and emerging mycotoxins, such as ochratoxin A, citrinin, moniliformin, and enniatins, to further increase the multiplexing capacity of the assay.
Methods
Chemicals and reagents
Mycotoxin standards—aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), aflatoxin G1 (AFG1), aflatoxin G2 (AFG2), aflatoxin M1 (AFM1), ochratoxin A (OTA), zearalenone (ZEA), T-2 toxin (T2), HT-2 toxin (HT-2), deoxynivalenol (DON), and fumonisin B1 (FB1) were purchased from Romer Labs (Biopure, Singapore). Goat-anti mouse (GAM) antibody (Code: KPL5210-0187) and rabbit anti-goat (RAG) antibody (Code: ab6697) were purchased from SeraCare (MA, USA) and Abcam (Cambridge, UK), respectively. All specific-mycotoxin antibodies and mycotoxin-conjugated bovine serum albumin (BSA) were obtained from Queen’s University Belfast (QUB), UK. Monoclonal antibodies against AFB1, T2, and DON were produced as described in detail by Oplatowska-Stachowiak et al.55 and Meneely et al.56,57 Mycotoxin-conjugated BSA was synthesized with slight modification56,58,59,60,61. Water and methanol (LC/MS grade) were supplied by Merck (Darmstadt, Germany). Other chemical reagents were purchased from Sigma-Aldrich (MO, USA).
Preparation of organic dye M424-labeled monoclonal antibody (GAM-M424 conjugation)
A luminescent organic dye named M424 was synthesized and linked to GAM antibody via a Schiff base-formation-reductive reaction as previously described by Charlermroj et al.21, with slight modifications. Briefly, 50 μL of 10 mg/mL M424 in dimethylsulfoxide (DMSO) was mixed with 500 μL of 1 mg/mL GAM antibody in phosphate buffer saline (PBS, 50 mM pH 7.4) before the introduction of 5.5 μL of 5 M cyanoborohydride in 1 N sodium hydroxide. After shaking at room temperature for 2 h, the mixture was added with 12 µL of ethanolamine (pH 6.6) to block any unreacted aldehyde sites of M424 and incubated at room temperature for 15 min with shaking. The supernatant containing a GAM-M424 conjugate was collected after centrifugation at 10,000 × g for 5 min and stored at 4 °C.
Fabrication of microarray lateral flow immunoassay strip (µLFIA)
The µLFIA strips were fabricated using a non-contact microarray dispenser (1520 CE) equipped with Biojet Elite™ dispenser (BioDot, Irvine, CA, USA) as previously described by Charlermroj et al.21. Mycotoxin-conjugated BSAs, including AFB1-BSA, T2-BSA, ZEA-BSA, DON-BSA, and FB1-BSA, were spotted on the microarray signal pad in quadruplicate with 500 μm in diameter with the following spotting parameters: 15 nL/spot, 200 μs open-time, 700-μm center-to-center spacing for both rows and columns and 60% relative humidity. The fluorescence signals from these four identical spots were averaged for further analysis. BSA and RAG antibodies were spotted in duplicate as negative and positive controls, respectively, while 0.15 mg/mL of M424 was used to mark the four corners of the µLFIA strip to facilitate spot localization for signal analysis. AFB1-BSA and BSA were spotted at 1 µg/mL, while the other mycotoxin-conjugated BSAs were spotted at 0.5 µg/mL. After spotting, each signal pad was dried at 37 °C for 2 h and immersed in 1 mL of optimized 10 mM borate buffer (pH 8.0) containing 0.5% polyvinyl pyrrolidone, 0.25% Triton X-100, and 1% BSA. Then, the signal pad was re-dried overnight at 37 °C. On a conjugate pad, a mixture of five monoclonal antibodies specific for target mycotoxins and M424-GAM antibody conjugate were spotted. To assemble a lateral flow strip, all the membrane pads (sample pad, conjugate pad, signal pad, and absorbent pads) were pasted onto a backing board (Fig. 5a) and cut into strips with widths of 4.0 mm using a guillotine cutter. Each strip was then inserted into a two-piece plastic cassette and stored at 4 °C.
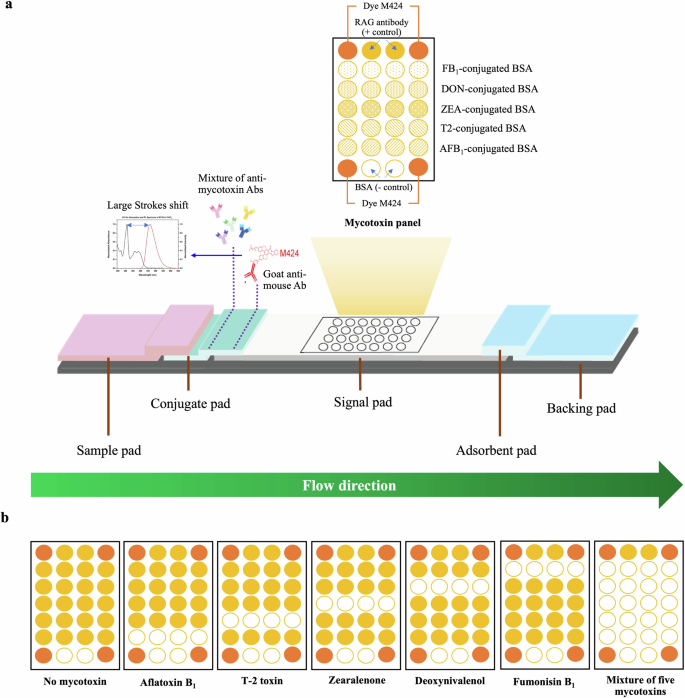
a µLFIA components and the spot layout of mycotoxin-bovine serum albumin (BSA), negative control, positive control, and dye M424 reference panel on a signal pad. b Expected results from the μLFIA strip using a sample with and without the target/relevant mycotoxins.
Microarray-strip reader
A cost-effective, rapid, and portable microarray reader was employed for on-site mycotoxin detection. The optical setup and signal correction of this reader, as reported in a previous study62, effectively compensate for fluorescence variation caused by intensity fluctuations and spatial non-uniformity in the excitation light. A light-emitting diode (LED: LST1-01G01-UV01-00, New Energy), in conjunction with an iris diaphragm (SM1D12, Thorlabs) and a bandpass filter (FF01-365/2-25, Semrock), generated a 365 nm wavelength light beam directed towards a μLFIA strip test using a dichroic beamsplitter (MD453, Thorlabs). The fluorescence emissions originating from the strip test were filtered by a 450 nm long-pass filter (FEL0450, Thorlabs) and then conveyed to an 8-bit camera (12.3 MP HQ, Raspberry Pi Foundation). Optomechanical components corresponding to these materials were created using a DLP 3D printer (Prusa SL1, Prusa Research). Using the Raspberry Pi platform (Raspberry Pi 4 model B, Raspberry Pi Foundation) programmed with Python 3, a microarray fluorescence image of the strip test with a resolution of 2592 × 1952 pixels was captured. The fluorescence signals from each spot, represented by the green channel, were extracted, while the corresponding background signals were obtained from the dark areas adjacent to those spots. An in-house software package for the reader was designed and developed to provide user-friendliness through a built-in 4.3-inch touchscreen (Cytron Technologies, Thailand). This software package comprised the backend for microarray-spot image acquisition and processing, mainly implemented using the OpenCV module, and the frontend, which offered a straightforward graphical user interface, using the PyQt module and Qt Designer.
Optimization of microarray lateral flow immunoassay multiplex detection conditions
In order to improve the robustness and sensitivity of the μLFIA, the following key parameters were optimized: concentrations of monoclonal antibodies, GAM-M424 conjugate, and running buffer components. More than 40 combinations of mixed anti-mycotoxin antibodies, ranging from 3 to 100 µg/mL, were investigated in this study. Briefly, 10 µL of anti-mycotoxin antibody mixtures at different concentrations were spotted on the conjugate pads. Then, 100 µL PBS containing 0.1% tween 20 and 0.5% BSA was introduced into the sample pad. After strip incubation, microarray spot signal intensities were captured and processed using the microarray reader to select the optimal concentrations of monoclonal antibodies.
In terms of GAM-M424 conjugation, different concentrations of the GAM-M424 conjugate (25, 75, 100, 110, 120, 130, 140, and 150 µg/mL) were tested to determine the optimal concentration that yielded the lowest background and highest signal intensity. Each GAM-M424 conjugate concentration and the optimized concentrations of anti-mycotoxin antibodies were fixed onto a conjugate pad. PBS containing 0.1% tween 20 and 0.5% BSA without target mycotoxins was used as a running buffer. After the experiments were completed, the test strips were incubated at 70 °C for 1 h. Microarray spot intensities on the strip tests were captured, processed, and analyzed.
Moreover, the running buffer of the developed method was optimized. Two detergents (tween 20 and Triton X-100) over a range of concentrations (0.1%, 0.2%, 0.5%, and 1%) were evaluated as a running buffer. In addition, six different molecular weights (MW) of PEG: MW100 (5%, 10%, and 20%), MW600 (5%, 10%, and 20%), MW3350 (5%, 7.5%, and 10%), MW6000 (0.5%, 1%, 2.5%, and 5%), MW8000 (1%, 2.5%, 3%, 4%, 5%, and 6%), and MW20,000 (1%, 1.5%, 2%, 2.5%, and 5%) were assessed. Optimized concentrations of mixed anti-mycotoxin antibodies and GAM-M424 conjugate were utilized for this experiment. Microarray spot images were captured and processed following strip incubation at room temperature.
Development of a green mycotoxin extraction solution for the lateral flow strip
A simple, rapid, and green sample preparation procedure was developed for the simultaneous extraction of five mycotoxins (AFB1, T2, ZEA, DON, and FB1). A wide range of surfactants and solutions were tested. PEG with a molecular weight of 20,000 (PEG 20K) was found to be compatible with the µLFIA strip and yield the highest recoveries of the target mycotoxins. Subsequently, six different concentrations (0.01%, 0.1%, 0.2%, 0.5%, 1%, and 2%) of PEG 20K supplemented with 20% ethanol (to improve mycotoxins recovery) were examined using LC–MS/MS to determine for the extraction capacity for target mycotoxins in rice. The recovery value was calculated as follows: %Recovery=(measured concentration of spiked sample/spiked concentration)×100. PEG 20K at 0.1% with 20% ethanol was found to give the highest mycotoxin recovery and was later used for further experiments, including optimization of sample extraction time and µLFIA assay validation.
Performance evaluation of microarray lateral flow immunoassay strip in rice sample
The specificity of µLFIA strip test for single and multiple mycotoxin detection was tested through a competitive assay format using both target and non-target mycotoxins, including AFB1, AFB2, AFG1, AFG2, AFM1, T2, HT-2, ZEA, DON, FB1, OTA, and different mycotoxin combinations (2-Plex, 3-Plex, 4-Plex, and 5-Plex).
In terms of sensitivity, a matrix-matched calibration curve was constructed for the quantitation of each target mycotoxin. The sensitivity and linearity of the assay were evaluated using the following mycotoxin concentration ranges: 0–2000 ng/mL for AFB1, ZEA, and T2 and 0–4000 ng/mL for DON and FB1. These concentration ranges were selected to cover the established MPL for mycotoxins in cereals and toxin levels commonly found in cereal samples in Asia19,21,24. The standard curves were plotted with the y-axis as log (%B/B0) and the x-axis as log (concentrations of mycotoxins). B is the signal intensity value of the mycotoxin-contaminated sample, and B0 is the signal intensity value of the blank sample. In addition, 20 blank (mycotoxin-free) samples were analyzed to estimate the limit of detection (LOD) and limit of quantification (LOQ) of the µLFIA. The LOD was calculated from a 3-fold of standard deviation obtained from the blank sample without any mycotoxin (n = 12), divided by the slope of a matrix-matched calibration curve, while LOQ was calculated from a 10-fold of standard deviation obtained from the blank sample without any mycotoxin (n = 12), divided by the slope of the matrix-matched calibration curve.
Moreover, the precision and accuracy of the assay were determined. Mycotoxin-free rice samples, confirmed by using a validated LC–MS/MS, were spiked at three different concentrations of multi-mycotoxin standards: low (L1), medium (L2), and high (L3) levels. The samples were mixed thoroughly and kept in a fume hood overnight to allow solvent evaporation. The final spiked concentration of each mycotoxin was 100, 200, and 500 µg/kg. These concentrations were also confirmed using a validated LC–MS/MS method described in the “Methods” section. The precision of the µLFIA was determined through intra-day (repeatability) and inter-day precisions (reproducibility). The intra-day precision was carried out by analysis of three replicates on the same day at three different concentration levels, while inter-day precision was evaluated by repeating the same procedure over three consecutive days. The data were used to calculate within-laboratory accuracy and precision and expressed as percentage relative standard deviation (%RSD). The recovery of each target compound was also calculated as the spot intensity ratio of blank samples spiked before and after sample extraction and expressed as a percentage. The µLFIA extraction recovery results were compared with the recovery results obtained using the validated LC–MS/MS method.
Test procedure for the microarray lateral flow immunoassay strip
The optimized assay conditions were used for the simultaneous analysis of five target mycotoxins in rice. Briefly, 1 g of contaminated rice samples was weighed into 15 mL propylene tubes. Subsequently, 5 mL of extraction solution containing 0.1% PEG 20K with 20% ethanol in 10 mM acetate buffer pH 5.0 was added to each tube. The tube was manually vortexed for 1 min and the mixture was filtered into a new Eppendorf tube using a 0.45 µm filter. The filtrate was diluted with an equal volume of running buffer solution containing 3.9% PEG 20K, 2% tween 20, and 1% BSA in 10 mM acetate buffer (pH 5.0) and briefly vortexed. An aliquot of 100 µL of the mixture was introduced into the sample pad of μLFIA strip. After incubation at room temperature for 15 min, semi-qualitative results from the strip were observed via the naked eye under ultraviolet (UV) light at 312 nm (ENB-260C/FE, New York, USA), while the quantitative results were obtained using the portable microarray reader. The total analysis time was ~17 min, and the sample-to-result workflow is illustrated in Fig. 6.
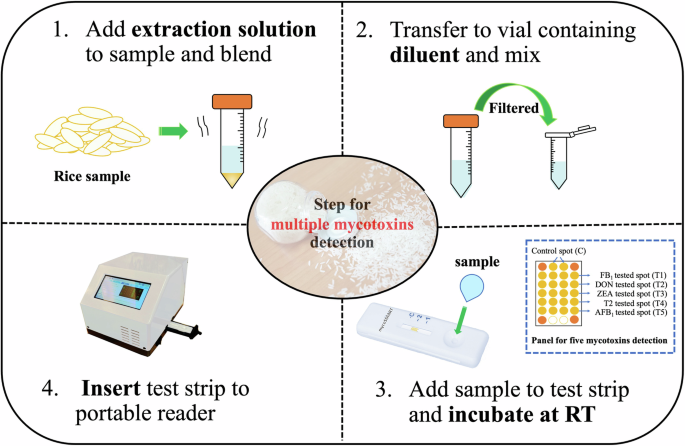
The developed and validated microarray lateral flow immunoassay strip with the green sample extraction enables the detection of multiple mycotoxins within 17 min using a portable microarray reader device.
To detect the mycotoxins, a direct competitive immunoassay approach was utilized due to their low molecular weights. Without any mycotoxin contamination, the specific monoclonal antibody fixed on the conjugate pad would bind to GAM-424 fluorescent organic dye before moving down the strip, and the antibody complex would be able to bind to the mycotoxin standard spots on the strip. With mycotoxin contamination, the antibody complex would bind to the mycotoxin present in the test sample, impeding it from binding to the standard mycotoxin-BSA spots on the strip (Fig. 5). As the specific antibodies are unable to react with mycotoxins on the signal pad, the signal intensity would be lower than that from the mycotoxin-free sample. The higher the level of mycotoxins in the test sample, the lower the spot signal intensity and vice versa (Fig. 5).
Sample preparation for LC–MS/MS analysis
One gram of blank (mycotoxin-free) and spiked rice samples were weighed into a 15 mL Eppendorf tube. Then, 5 mL of 70% methanol at pH 3 was added. The tube was vortexed for 30 min and centrifuged at 100 rpm for 10 min. An aliquot of the supernatant was transferred into a new Eppendorf tube and diluted with an equal volume of acetonitrile with 2% acetic acid. The mixture was vortexed and filtered through a 0.22 µm PTFE filter into a glass vial for LC–MS/MS analysis.
The qualitative and quantitative determination of mycotoxins were carried out on an Agilent 1260 Infinity II HPLC system coupled to an Ultivo triple quadrupole mass spectrometry, equipped with a jet stream electrospray ionization source (LC–MS/MS). The analytes were separated through gradient elution on Gemini 5 μm C18 110 Å 50 × 4.6 mm i.d. column maintained at 30 °C. The mobile phases consisted of water (A) and methanol (B), both containing 0.1% formic acid and 5 mM ammonium hydroxide. The following gradient elution protocol was followed for mycotoxin analysis: 0–2 min (60% A), 2–4 min (50% A), 4–6 min (20% A), and 6–9 min (60% A). The total run time was 9 min with a flow rate of 0.9 mL/min and sample injection volume of 2 μL.
The mass spectrometer (MS) was operated in dynamic multiple reaction monitoring mode (dMRM), filtering one precursor and two product ions of each mycotoxin. The MS parameters, such as collision energy and fragmentor voltage, and ion transitions, were optimized for each compound. The ion source was operated in both positive and negative modes, with a gas temperature of 250 °C, a gas flow of 10 L/min, a nebulizer pressure of 10 psi, and a capillary voltage of 4 kV or −4 kV. Data acquisition and analyses were performed using the Agilent Mass Hunter software package, which includes acquisition for Ultivo (Ver 1.1.2222), Optimizer (Ver 1.1.2222), quantitative analysis (Ver 10.0.707.0), and qualitative analysis (Ver 10.0.10305.0).
Statistical analysis
Statistical analysis was carried out using SPSS version 26.0 for Windows, and significant differences (p < 0.05) between means were determined by Duncan’s multiple range test.


Responses