Antimicrobial resistant enteric bacteria are widely distributed among environmental water sources in Dhaka, Bangladesh

Introduction
Antimicrobial-resistant (AMR) and multidrug-resistant (MDR) bacteria pose a major threat to global public health, exacerbated by the environmental dissemination of resistant isolates1. In 2019, AMR was directly responsible for 1.27 million deaths and associated with an estimated 4.95 million deaths globally2. Misuse of antibiotics in humans, both in hospitals, communities, and in animals including livestock, poultry, and aquaculture, contributes to the introduction of antibiotics and antibiotic-resistant genes (ARGs) into soil and water bodies3. By 2050, the estimated use of antimicrobials in humans and animals will be increased by about 70–80% in low- and middle-income countries (LMICs)4,5. The generation of antibiotic resistance genes through natural selection can be amplified by the estimated concentrations of antibiotics present in the environment, transforming the environment into a significant reservoir for the continued spread and multiplication of ARGs3,4,5,6,7,8,9.
Surface water pollution in LMICs raises considerable concerns. Both treated and untreated wastewater discharged into rivers and streams has been contributing to potential outbreaks of waterborne infections, which may act as a significant mechanism for spreading antibiotic resistance genes among pathogenic bacterial populations10,11. Reservoirs of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) are present in wastewater, potentially facilitating the accelerated horizontal transfer of resistance determinants among native microbial communities12,13. Consequently, the detection of ARBs and ARGs in environmental samples has been directly linked to the rise of antibiotic resistance in clinical infection treatments4,7,12,13,14,15,16,17. We performed a comprehensive study to determine the presence of ARBs and ARGs and the role of the resistance genes carrying isolates in disseminating any of the resistance genes.
Consistent with the One-Health concept, which recognizes the interdependence of human, animal, and environmental health, a comprehensive environmental study of pathogenic and antibiotic-resistant bacteria serves as an effective method to evaluate and infer the prevalence of ARB and resistance genes within human communities3,7,11,12,13,18. In Bangladesh, the health infrastructure is significantly strained by the prevalence of diarrheal diseases, a situation aggravated by the resistance of causative entero-pathogens to antibiotics19,20. Mixing of contaminated water during seasonal floods and improper water treatment serves as one of the major sources for transmitting these entero-pathogens to humans, hence, elucidating the antimicrobial resistance profiles of these bacterial isolates within environmental water sources is paramount21.
Prior research has often been limited in scope, typically concentrating on a single water source, such as hospital, industrial, pharmaceutical wastewater, or surface water22,23,24. There remains a gap in extensive and comparative research across all potential environmental water sources regarding the multidrug resistance of enteric bacteria and the evolutionary relation of these resistance genes. Therefore, this study was executed to investigate the antimicrobial resistance (AMR) and multidrug-resistance (MDR) profiles of enteric bacterial isolates, particularly E. coli, V. cholerae, Salmonella spp., and Shigella spp., and involvement of resistance gene carrying isolates in the dissemination of AMR. Outcomes from this study are anticipated to contribute new perspectives on environmental sources of AMR, informing strategic policy development to address and curb the substantial health implications associated with the use of antibiotics and sources of resistant bacteria in resource-limited communities. The main aim of this study was to investigate the comparative distribution of antimicrobial-resistant and multidrug-resistant enteric bacteria, their resistance genes, and the molecular association of resistance isolates in different water sources.
Results
Total viable and total gram-negative count
The total viable count on the nutrient agar plate ranged between 1.7 × 103 cfu/ml and 2.1 × 109 cfu/ml and the highest count was found in the sample of a canal located in Bank Town (2.1 × 109 cfu/ml) and the lowest count was found from the sample of irrigation water in the Gerua area near Jahangirnagar University (1.7 × 103 cfu/ml). Gram-negative bacterial count on the MacConkey agar plate ranged between 1 × 102 cfu/ml and 1.52 × 106 cfu/ml and the highest count was found in the sample of a canal located in Bank Town (1.52 × 106 cfu/ml) and the lowest count was found from the sample of irrigation water in the Gerua area near Jahangirnagar University (1 × 102 cfu/ml). Sampling regions were indicated on the map with legends focusing on different sources (Fig. 1).
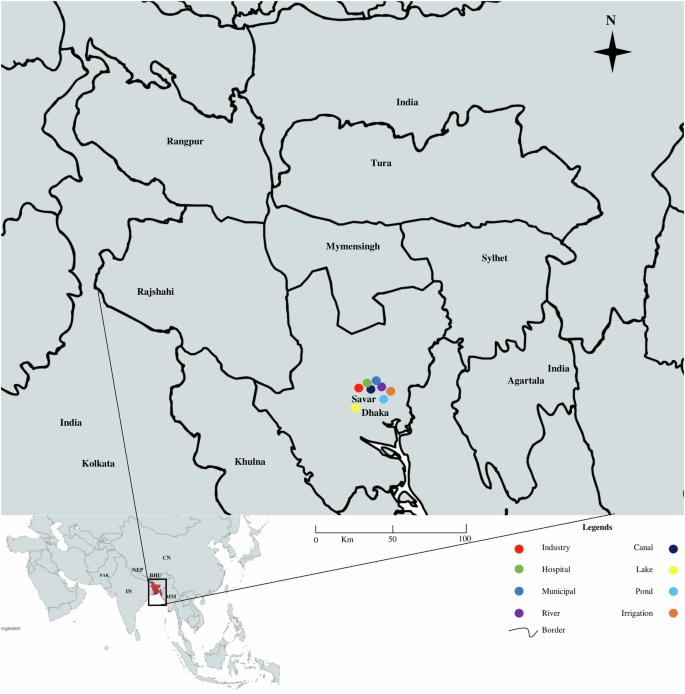
The map was created using the ArcGIS (USA) software.
Association of physicochemical parameters
Observations revealed significant fluctuation in the temperature of the water samples, ranging between 16.3 and 30.5 °C. The pH levels of these samples also varied significantly, ranging from pH 5.3–8.8 (Supplementary Table 1). The dissolved oxygen (DO) levels varied across different water samples. Industrial wastewaters exhibited the lowest DO concentrations, with values spanning from 1.5 to 4.5 mg/L. Hospital and municipal wastewaters displayed DO ranges of 2.0–4.8 mg/L and 2.6–5.0 mg/L, respectively. Industrial wastewaters were associated with the most elevated biological oxygen demand (BOD) values, which fluctuated from 200 to 450 mg/L. Hospital wastewaters and municipal wastewaters had BOD values in the ranges of 100–250 mg/L and 200–350 mg/L, respectively. Further, chemical oxygen demand (COD) varied from 25.05 mg/L to 442.31 mg/L among different sources.
We determined the association of the prevalence of isolates with physicochemical factors among different sources of water. A significant association between prevalence and temperature was found in industrial effluent (p = 0.04), municipal wastewater (p = 0.01), river water (p = 0.03), lake water (p = 0.01), pond water (p = 0.005) and irrigation water (p = 0.01) (Table 1). However, the pH of the river (p = 0.01), canal (p = 0.04), and lake water (p < 0.001) were significantly associated with bacterial prevalence. Further, we found a significant association (p ≤ 0.05) of the prevalence of enteric bacteria with DO, BOD, and COD in the majority of water sources.
Propionate frequency of enteric bacteria in water sources
Among the bacterial isolates detected, E. coli was the most prevalent (29.68%, 46 of 155) followed by Vibrio cholerae (27.74%, 43 of 155), Shigella spp. (22.58%, 35 of 155), and Salmonella spp. (20.00%, 31 of 155), respectively (Supplementary Fig. 1 part A).
The highest frequency of bacterial isolates was found in hospital wastewater (26%-31%), followed by municipal wastewater (15–23%), canal water (13–20%), and industrial effluent (11–17%), respectively. The lowest frequency of bacterial isolates was detected in Lake water (0–4%), followed by irrigation water (2–3%), pond water (3–8%), and river water (9–13%), respectively (Supplementary Fig. 1 part B).
Resistant bacteria are widely distributed in water sources
Partial amplicons of 16S rRNA and specific resistant genes were used to determine the species of resistance gene-carrying isolates. The detailed protocol is previously published elsewhere. Multiple sequence alignment (MSA) was conducted by using ClustalW and trees were built by using the maximum likelihood method following 1000 bootstrap values. We randomly selected 18 amplicons of E coli, 18 amplicons of V. cholerae, 16 amplicons of Salmonella spp., and 16 amplicons of Shigella spp. from both phenotype and genotype-resistant isolates for phylogenetic analysis. These 16S rRNA-based trees showed that circulating isolates of enteric bacteria in Bangladesh during recent times are highly similar across different sources. Isolates of E coli and V. cholerae from hospital wastewater and industrial effluent were closely related to isolates from lakes, rivers, and pond water (> 99.8% similarity; <0.002 divergence) (Fig. 2A, B). Further, isolates of Salmonella spp., and Shigella spp. from ponds, rivers, and canals clustered closely with the isolates of municipality and hospital wastewater (> 99.5% similarity; <0.005 divergence) (Fig. 3A, B).
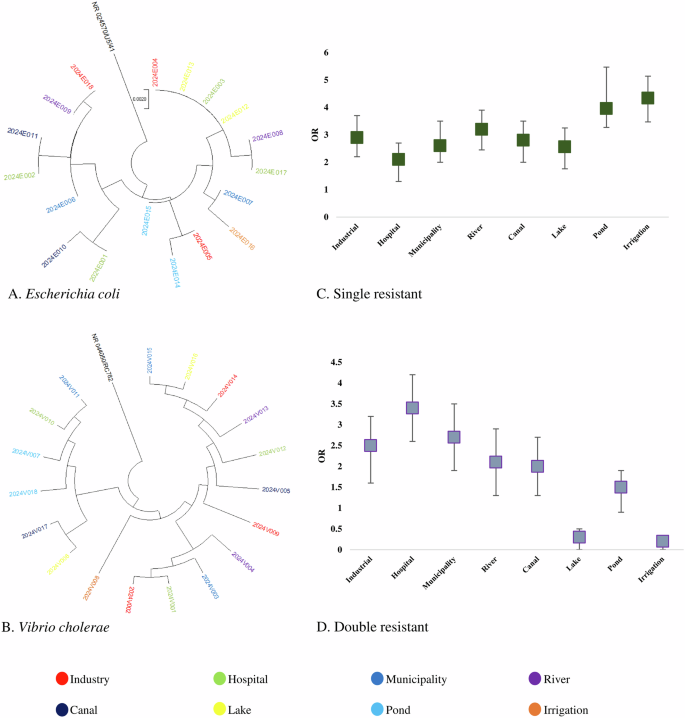
Phylogenetic tree of antimicrobial-resistant (A) Escherichia coli, (B) Vibrio cholerae isolates and risk of prevalence of antibiotic-resistant. (C) Single-resistant isolates and D Double-resistant isolates.
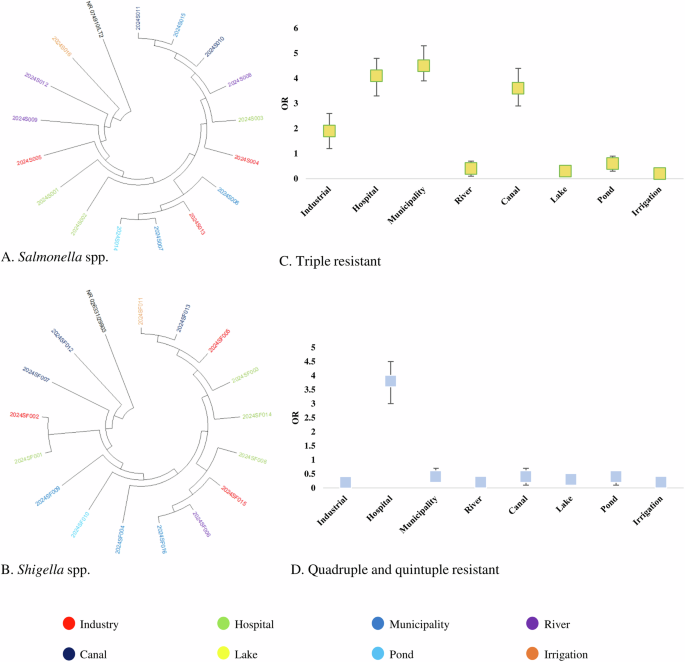
Phylogenetic tree of antimicrobial-resistant. A Salmonella spp., and (B) Shigella spp. isolates and risk of prevalence of antibiotic-resistant. C Triple-resistant isolates and (D) quadruple and quintuple-resistant isolates.
We used 40 amplicons of antibiotic-resistant genes including blaTEM, qnrB, tetA, mcr-1, and sul-1. We found that resistant genes from environmental sources were closely related to widely distributed resistant genes in enteric bacteria isolated from animals and humans in recent times in Bangladesh. Our study-resistant genes including blaTEM, qnrB, tetA, mcr-1, and sul-1 were highly similar (> 99.5% similarity; <0.002 divergences) with previously reported resistant genes isolated from clinical and environmental strains (Fig. 4).
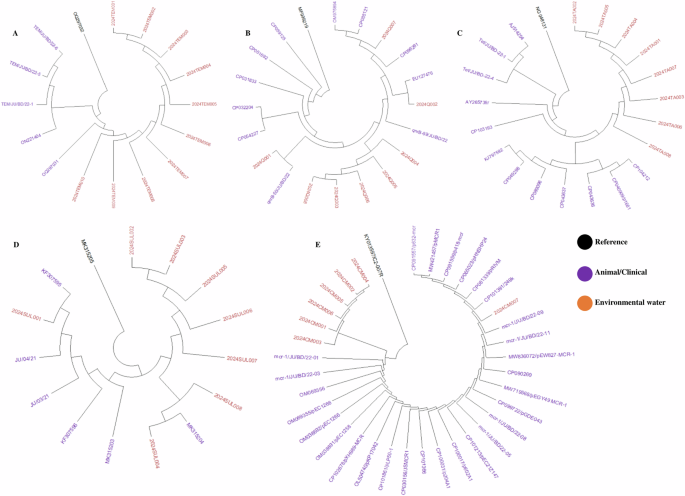
Phylogenetic tree of antimicrobial resistant marker of (A) Beta-lactam, (B) Quinolone (C) Tetracycline, (D) Cotrimoxazole, and (E) Colistin among the isolates of freshwater samples.
Association of MDR isolates with the sources of isolation
Odds of prevalence of single resistant bacteria were higher in irrigation water (OR: 4.34, 95% CI: 3.56–4.98), followed by pond water (OR: 3.96, 95% CI: 3.18–4.85), river water (OR: 3.21, 95% CI: 2.38–3.92), industrial effluent (OR: 2.91, 95% CI: 2.21–3.72) and canal water (OR: 2.83, 95% CI: 1.98–3.61), respectively (Fig. 2C). We found significantly higher odds of prevalence of double resistant bacteria in hospital wastewater (OR: 3.43, 95% CI: 2.68–4.29), followed by municipality wastewater (OR: 2.72, 95% CI: 1.95–3.63), industrial effluent (OR: 2.52, 95% CI: 1.68–3.34) and river water (OR: 2.17, 95% CI: 1.46–3.06), respectively (Fig. 2D). Higher odds of triple resistance were prevalent among bacteria in municipality wastewater (OR: 4.53, 95% CI: 4.13–5.25), hospital wastewater (OR: 4.16, 95% CI: 3.57–4.98), canal water (OR: 3.65, 95% CI: 3.11–4.26) and industrial effluent (OR: 1.93, 95% CI: 1.15–2.72) (Fig. 3C). Further, we found higher odds of quadruple and quintuple resistance only in hospital wastewater (OR: 3.86, 95% CI: 3.17–4.61) (Fig. 3D). The odds of prevalence of MDR bacteria were significantly lower ( < 1) in rivers, lakes, ponds, and irrigation water (Fig. 2C, D).
Prevalence of AMR isolates associated with sources
Multivariable logistic models were used and clustered at the source level to assess the source factors associated with the detection of phenotypic antimicrobial-resistant isolates. Primary outcomes included being resistant to an antibiotic(s) (1) or being sensitive (0). Models were defined for all ten resistance phenotypes. The findings were restricted to only statistically significant variables. Odds above 1 indicate increased odds of resistance in specific sources and <1 decreased odds of isolates with phenotypic resistance.
Higher odds of ceftriaxone-resistant phenotypes were present among enteric bacteria circulating in hospital wastewater (OR: 3.21, 95% CI: 2.36–3.95), followed by industrial effluent (OR: 2.15, 95% CI: 1.66–2.38), and municipal wastewater (OR: 1.64, 95% CI: 0.92–2.48), respectively. Significantly higher odds of imipenem-resistant isolates were only found in hospital wastewater (OR: 5.34, 95% CI: 4.48–5.97) (Fig. 5). Erythromycin-resistant bacteria were circulating in the majority of the water sources. However, the highest odds of prevalence were found in hospital wastewater (OR: 2.93, 95% CI: 2.15–3.76), followed by industrial effluent (OR: 2.32, 95% CI: 1.51–3.13). Isolates in hospital wastewater had higher odds of phenotypic resistance to tetracycline (OR: 2.53, 95% CI: 1.94–3.13), doxycycline (OR: 2.91, 95% CI: 2.15–3.74), chloramphenicol (OR: 3.15, 95% CI: 2.28–3.78), colistin (OR: 3.94, 95% CI: 3.08–4.63) and cotrimoxazole (OR: 3.81, 95% CI: 2.97–4.51) than any other sources (Fig. 5). Bacterial isolates from industrial effluent had the highest odds of resistance against ciprofloxacin (OR: 2.72, 95% CI: 2.97–4.51) and norfloxacin (OR: 2.84, 95% CI: 2.97–4.51). Further, higher odds of prevalence of norfloxacin and ciprofloxacin-resistant bacteria were found in municipal wastewater (OR: 2.31, 95% CI: 1.56–3.03 and OR: 2.14, 95% CI: 1.47–2.75) and river water (OR: 2.16, 95% CI: 1.43–2.67) (Fig. 5).
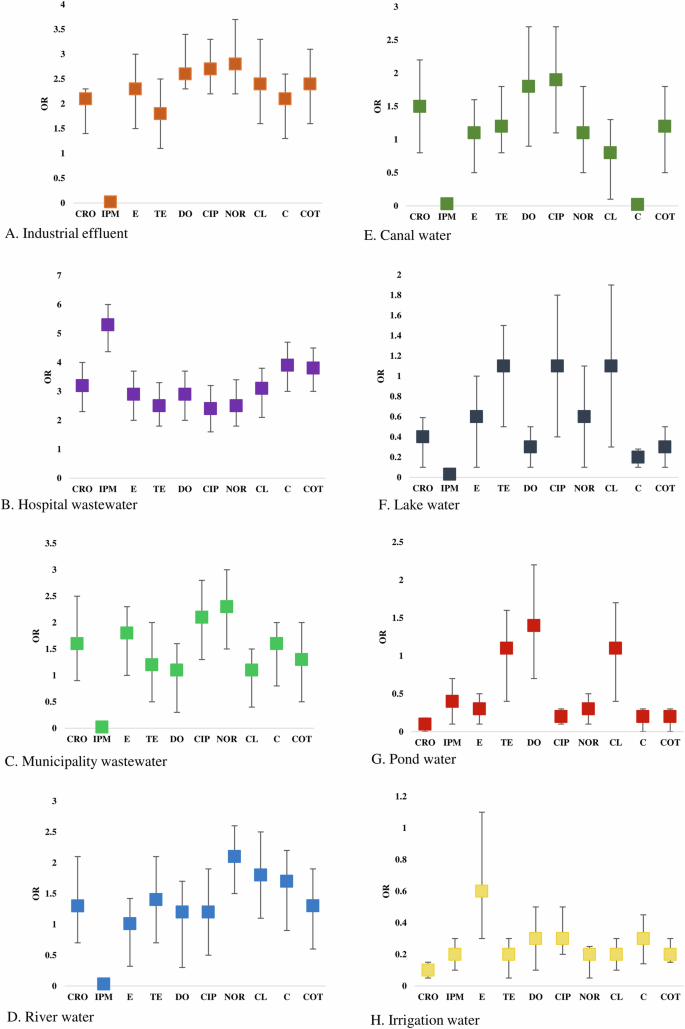
Risk assessment of the prevalence of phenotypic resistant bacteria in (A) Industrial effluent, (B) Hospital wastewater, (C) Municipality wastewater, (D) River water, (E) Canal water, (F) Lake water, (G) Pond water, (H) Irrigation water.
Antimicrobial resistance pattern of the enteric bacteria
Among the isolates of E. coli, the highest frequency of resistance was observed to ceftriaxone (46.65%, 21 of 46) and erythromycin (46.65%, 21 of 46), and followed by tetracycline (26.09%, 12 of 46), and chloramphenicol (23.91%, 11 of 46), doxycycline (22%, 10 of 46), ciprofloxacin (22%, 10 of 46) and norfloxacin (20%, 9 of 46), respectively. Conversely, isolates of E. coli showed the highest frequency of sensitivity to imipenem (95.65%, 44 of 46), and colistin (78.26%, 36 of 46) (Table 2). Isolates of V. cholerae were less resistant than the isolates of E. coli against all the tested antibiotics. The highest percentage of resistance was found to erythromycin (41.86%, 18 of 43), followed by ceftriaxone (35%, 15 of 43), tetracycline (23%, 10 of 43), doxycycline (19%, 8 of 43), ciprofloxacin (14%, 6 of 43) and norfloxacin (14%, 6 of 43), respectively (Table 2).
The majority of the isolates of Salmonella spp. were resistant against ceftriaxone (35.48%, 11 of 31), followed by erythromycin (29%, 9 of 31), tetracycline (23%, 7 of 31), and ciprofloxacin (19%, 6 of 31), respectively, while higher frequency of sensitivity was found against colistin and (90.32%, 28 of 31) and imipenem (96.77%, 30 of 31). Similarly, the isolates of Shigella spp. exhibited a higher frequency of resistance against ceftriaxone (42.86%, 15 of 35), erythromycin (40%, 14 of 35), and tetracycline (28.57, 10 of 35) (Table 2).
Comparative antibiotic resistance profile of the isolates
Among the enteric bacteria, E. coli isolated from hospital wastewater showed the highest frequency (35%, 95% CI 20–100%) of phenotypic resistant followed by industrial effluent (18.5%, 95% CI 0–25%), municipal wastewater (14.3%, 95% CI 0–22%), river water (13.3%, 95% CI 0–22%), and canal water (10.9%, 95% CI 0–20%), respectively against the tested antibiotics. Isolates of E. coli in hospital wastewater were most resistant against imipenem (100%), followed by colistin (40%), cotrimoxazole (37.5%), doxycycline (30%), and erythromycin (28.6%), respectively (Fig. 6A).
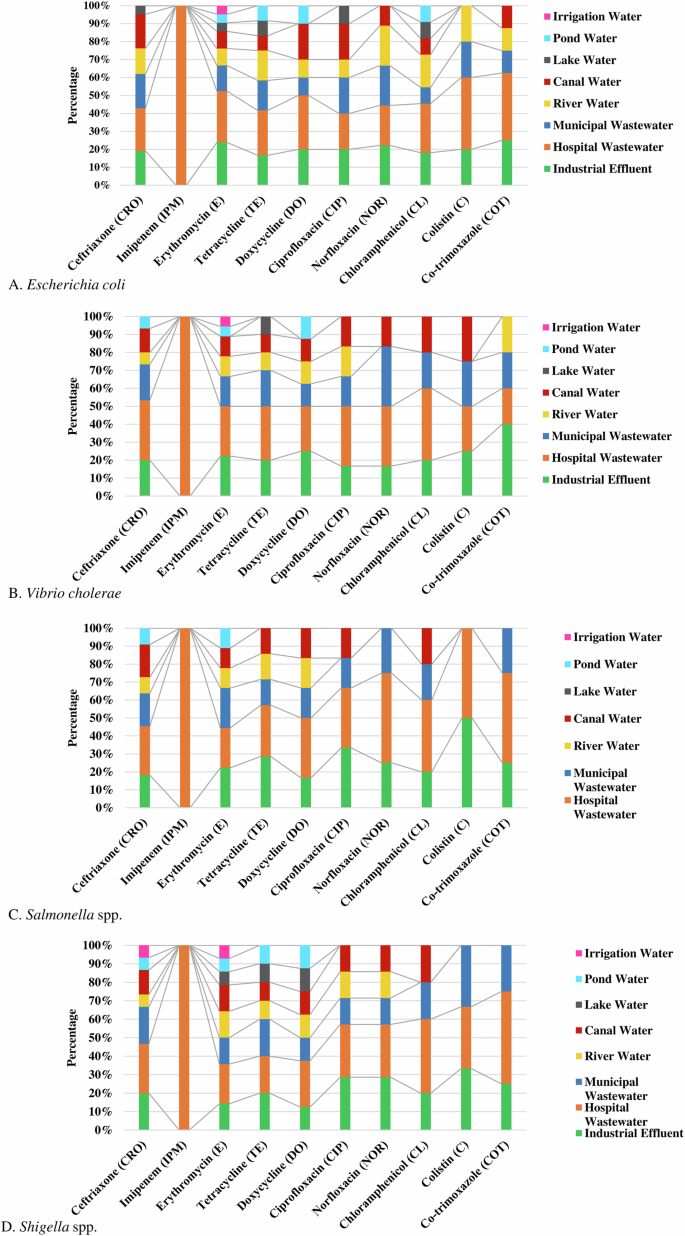
Source distribution of antibiotic-resistant A Escherichia coli, (B) Vibrio cholerae, (C) Salmonella spp., and (D) Shigella spp.
Isolates of V. cholerae from hospital wastewater (37%, 95% CI 20–100%) showed the highest frequency of resistance followed by industrial effluent (20.5%, 95% CI 0–40%), municipal wastewater (18.4%, 95% CI 0–33%) and canal water (12.5%, 95% CI 0–25%), respectively. Significant frequency of V. cholerae in hospital wastewater and industrial effluent were resistant against chloramphenicol (40%) and colistin (40%) (Fig. 6B).
Salmonella spp. in hospital wastewater (43.5%, 95% CI 22–100%) showed the highest frequency of resistance, followed by industrial effluent (23.9%, 95% CI 0–50%), and municipal wastewater (15.8%, 95% CI 0–25%), respectively. About 50% of the isolates of Salmonella spp. in hospital wastewater were resistant to norfloxacin, cotrimoxazole, and colistin (Fig. 6C). Similarly, a higher frequency of resistant Shigella spp. was isolated from hospital wastewater (37.5%, 95% CI 20–100%), industrial effluent (20.3%, 95% CI 0–33%), and municipal wastewater (17.3%, 95% CI 0–33%), respectively (Fig. 6D).
About 100% of the enteric bacteria in hospital wastewater were resistant to imipenem, while bacterial isolates (100%) from all other water sources were highly sensitive to imipenem (Fig. 6). Isolates of enteric bacteria resistant against colistin were commonly found in hospital wastewater (37%, 95% CI 25–50%), industrial effluent (32%, 95% CI 20–50%), municipal wastewater (19.5%, 95% CI 0–33%). Resistant isolates against erythromycin were widely distributed in the majority (95%) of the water sources. The majority of the enteric bacteria (90%) in pond, lake, and irrigation water were sensitive to the tested antibiotics (Fig. 6).
Prevalence of multidrug resistance enteric bacteria
Multidrug resistance (MDR) was defined as phenotypic or genotypic resistance of a single isolate to more than three groups of antibiotics. We found MDR isolates against cephem (ceftriaxone), macrolides (erythromycin), tetracyclines (tetracycline), cotrimoxazole and phenicol (chloramphenicol).
We found about 60% of enteric bacteria were resistant to a single group of antibiotics, followed by double (26%), triple (10%), quadruple (3%) and quintuple (1%) resistant, respectively (Fig. 7A). Overall, MDR in phenotype was found among 14% of bacteria. Phenotypic resistance against two groups of antibiotics was detected at the highest frequency in Shigella spp. (29%), followed by V. cholerae (28%), Salmonella spp. (26%) and E. coli (22%), respectively. Multidrug resistance including triple, quadruple, and quintuple resistance was found in highest prevalence in E. coli (15%), V. cholerae (15%), followed by Salmonella spp. (13%) and Shigella spp. (11%), respectively (Fig. 7).
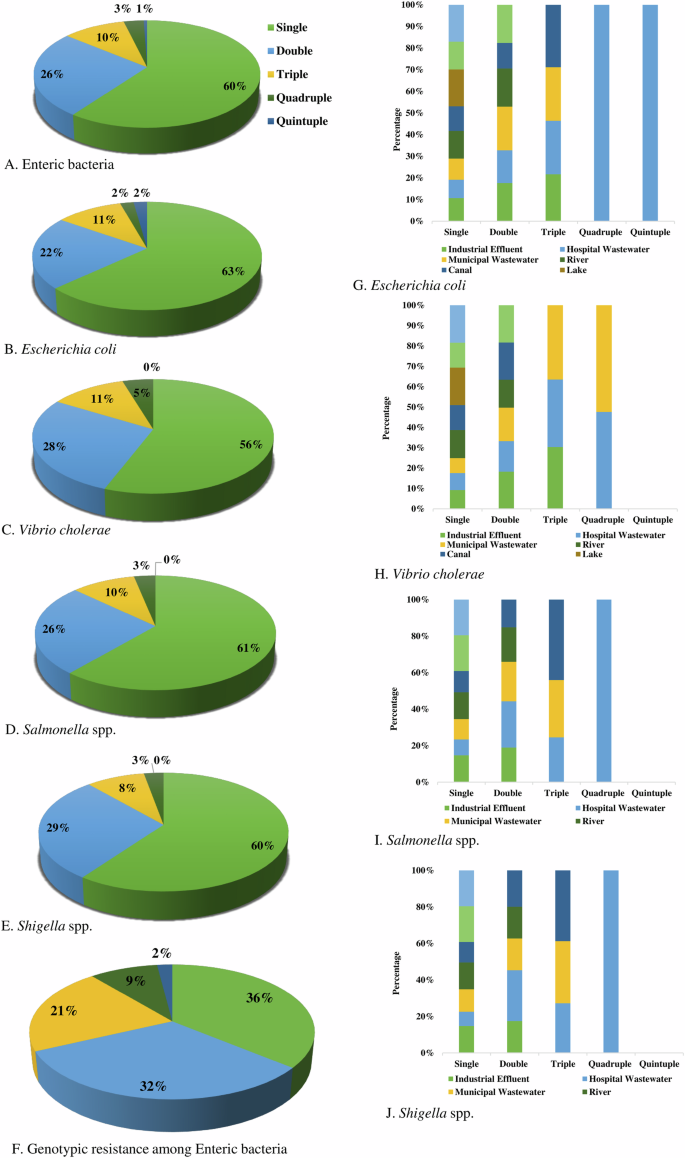
Proportionate frequency of multidrug-resistant phenotype in (A) Enteric bacteria, (B) Escherichia coli, (C) Vibrio cholerae, (D) Salmonella spp., (E) Shigella spp. and (F). Genotypic resistant isolates, and comparative frequency of multidrug-resistant (G). Escherichia coli, (H) Vibrio cholerae, (I) Salmonella spp., and (J) Shigella spp. in different water sources.
The prevalence of genotypic resistance among bacterial isolates was determined by the presence of specific resistant genes. Single resistance (36%) was most frequent followed by double (32%), triple (21%), quadruple (9%), and quintuple (2%), respectively (Fig. 7F).
Distribution of MDR isolates in water sources
Multidrug-resistant E. coli isolates were most prevalent in hospital wastewater, with approximately 7.14% exhibiting both quadruple and quintuple resistance, and about 14.29% showing triple resistance. In canal water, around 16.67% of the isolates showed triple resistance, while in municipal wastewater, nearly 14.29% of isolates displayed triple resistance. In industrial effluent, almost 12.50% of isolates exhibited triple resistance (Fig. 7G).
Multidrug-resistant V. cholerae isolates were most prevalent in municipal and hospital wastewater. In municipal wastewater, approximately 20% of isolates showed triple resistance, while about 10% exhibited quadruple resistance. In hospital wastewater, around 18.18% of isolates demonstrated triple resistance, and nearly 9.09% displayed quadruple resistance. Approximately 27% of isolates in industrial effluent showed triple resistance (Fig. 7H).
Multidrug-resistant Salmonella spp. isolates were most prevalent in hospital wastewater, with about 11.11% exhibiting both triple and quadruple resistance. In canal water, around 20% of isolates showed triple resistance, while approximately 14% of isolates in municipal wastewater exhibited triple resistance (Fig. 7I).
Multidrug-resistance Shigella spp. isolates were most prevalent in hospital wastewater, with about 10% exhibiting both triple and quadruple resistance. In canal water, around 14.29% of isolates showed triple resistance, while approximately 12.50% of isolates in municipal wastewater exhibited triple resistance (Fig. 7J).
Association of phenotypic and genotypic resistance
The prevalence and association of phenotypic and genotypic resistance among the enteric bacterial isolates were evaluated. We found resistant genotype against quinolones (qnr genes), β-Lactams (blaTEM, blaCTX-M-15, blaOXA), and antagonists of the folate pathway (co-trimoxazole-sul-1 genes), tetracycline and colistin. The highest prevalence of resistant genotype was found against β-lactams (30%-37%), followed by ciprofloxacin (23%-31%), tetracycline (13%-24%), cotrimoxazole (11%-13%), and colistin (3%-10%), respectively (Table 3).
We wanted to determine if the presence of these resistance genes was associated with phenotypic resistance among these bacterial isolates. We found a significant association between the prevalence of genotype and phenotype resistance among the enteric isolates. A significant association was observed between phenotypic and genotypic resistance among the isolates of E. coli for ceftriaxone (p = 0.04), ciprofloxacin (p = 0.002), tetracycline (p = 0.005), cotrimoxazole (p = 0.001) and colistin (p = 0.005). Similarly, among the isolates of V. cholerae, Salmonella spp., and Shigella spp., the association of phenotypic and genotypic resistance was statistically significant (p value ≤ 0.05) (Table 3).
Discussion
Ecological habitats, especially water bodies connected to hospital waste, industrial waste, and municipal sewage systems, become heavily contaminated with antimicrobial agents and resistant bacteria3,4,5,6,7,8,10,11,12,13. These bacteria often carry mobile resistance gene cassettes, which facilitate the exchange of genetic material and the emergence of new clinically significant resistant bacteria12,13,14. The Savar region has approximately 54 hospitals and clinics, 20 diagnostic services, 31 pharmaceutical industries with antimicrobial production units, and dairy and animal farms. The inadequacy of local wastewater treatment systems results in the discharge of untreated effluents containing active antimicrobial agents and antibiotic-resistant bacteria23,24. These water sources with antimicrobial-resistance bacteria (ARBs) may be involved with direct and cross-transmission of AMR among humans, animals, and the environment. Seasonal floods pave the way for mixing water from different water bodies in localities like Savar. Contaminated water is a primary medium for transmitting these pathogens to humans, highlighting the importance of understanding the spread of antimicrobial-resistant (AMR) bacteria in environmental water sources11,12,13,22,23,24,25,26.
We evaluated the distribution of antimicrobial-resistant enteric bacteria in water in central Bangladesh, examined the contribution of these sources in containing AMR and multidrug-resistant (MDR) bacteria, and explored the comparative evolutionary relationship of these isolates. Our study highlighted six major points. First, the prevalence of enteric bacteria in these water sources was associated with different physicochemical factors including pH, temperature, BOD, and COD. The seasonal changes in the count of enteric bacteria were correlated to the temperature and pH of water bodies. The prevalence of AMR isolates was relatively higher in months with higher average temperatures. A previous study in the USA also reported an increase in temperature of 10 °C might contribute to a 4.2%, increase in resistance in Escherichia coli isolates27. Though our findings resonate with previous findings, the roles of environmental factors in AMR should be explored in detail in the future. Second, the isolated enteric bacteria in water in central Bangladesh showed a high prevalence of antimicrobial resistance isolates of E. coli (44%), V. cholerae (35%), Shigella spp. (34%), and Salmonella spp. (27%). Isolates from industrial effluent, hospital wastewater, municipal wastewater, river water, pond water, lake water, canal water, and irrigation water showed antimicrobial resistance to at least one antibiotic. Approximately 40% of the isolates, of E. coli, V. cholerae, Salmonella spp., and Shigella spp., showed higher resistance to ceftriaxone, erythromycin, norfloxacin, ciprofloxacin, and tetracycline. This evidence is corroborated by prior research conducted in environmental water samples and clinical samples in Bangladesh and other developing and developed countries1,7,22,23,24,25,26,27,28,29,30. Further, MDR phenotype and genotype were commonly detected against beta-lactam, tetracycline, erythromycin, cotrimoxazole, and chloramphenicol in enteric bacteria in this study. We have found a relatively higher frequency of AMR isolates in environmental water samples compared to previous studies in Bangladesh22,23,24,25,26. Approximately 50% of the E. coli and Shigella spp. isolates, and around 33.33% of the V. cholerae and Salmonella spp. isolates demonstrated resistance to ceftriaxone and erythromycin. About 25% of the E. coli, Shigella spp., and V. cholerae isolates were resistant to tetracycline and doxycycline. The presence of AMR isolates of Enterobacteriaceae in freshwater has been reported from countries worldwide including the USA, Japan, India, China, Brazil, Switzerland, Portugal, Nepal, France, and Finland6,7,11,27,28,29,30,31,32. Apart from the antibiotic selection pressure and contamination of different water bodies with resistant isolates, bacteria are gaining the properties of AMR by mutations and co-selection and making a pool of resistome in environmental habitats. However, the risk of easily transmitting ARBs from freshwater to humans is higher in LMICs than in developed countries. These findings call for more vigilant monitoring of the water system and the adoption of a proper policy to ensure safe water for everyone in developing countries like Bangladesh.
Consistent with AMR isolates, we found a higher prevalence of MDR Enterobacteriaceae from these freshwater sources. The presence of MDR isolates has been reported previously from surface water in Bangladesh, India, and China. However, we found a higher number of MDR isolates in environmental water than any previous reports in the developed countries11,32. The profiling of multidrug resistance (MDR) among the isolated bacteria was concerning. The highest frequency of multidrug resistance was found among the isolates of V. cholerae (16%), followed by E. coli (15%), Salmonella spp. (13%), and Shigella spp. (11%). Notably, 2.17% of E. coli isolates demonstrated quintuple resistance, while quintuple resistance was absent in other bacterial isolates. Among E. coli isolates, 10.87% and 2.17% isolates demonstrated triple and quadruple resistance, respectively, which is in contrast with previous studies23,24. Among the isolates of Salmonella spp., 9.68% showed triple resistance, while 3.22% showed quadruple resistance. This pattern partially aligns with the findings in Thailand32. For Shigella spp., 8.57% and 2.86% of isolates demonstrated triple and quadruple resistance, respectively, which contrasts with previous studies in Bangladesh22,23,24. While comparing these findings with previous studies in other countries, we found it alarming that the number of isolates with diverse resistance mechanisms is increasing in environmental freshwater in Bangladesh. There may be several reasons behind this alarming increase of the MDR Enterobacteriaceae. Firstly, the rate of successful horizontal transfer of AMR genes might have increased. Secondly, the isolates have become more competent in expressing these resistance properties11. Lastly, the AMR isolates are frequently coming near each other to share AMR genes in environmental sites like freshwater bodies.
Third, the presence of V. cholerae, Salmonella spp., and Shigella spp. in different water sources indicated probable contamination of these water by humans and animals. This finding predicted that potential enteric pathogens are regularly contaminating water in central Bangladesh and becoming an environmental hub for the exchange of AMR genotypes among them. These results concur with earlier research conducted in Bangladesh, China, India, Ghana, Nigeria and Thailand22,23,24,25,26,27,28,29,30,31,32. A previous meta-study also found that the wastewater from hospitals, pharmaceutical industries, and municipality areas was harboring higher concentrations of antibiotics and a higher number of AMR isolates in several developed and developing countries11,32. Similarly, we have found that the wastewater from these sources contributes to higher enteric pathogens and AMR isolates in different freshwater sources in Bangladesh.
Fourth, a comparison of MSA data and phylogenetic analysis demonstrated that isolates of E. coli, V. cholerae, Salmonella spp., and Shigella spp. from our samples are genetically diverse and widely distributed across different sources. The divergence was minimal among the isolates of E. coli in these sources. The high similarity of environmental isolates in this study was found with isolates from animals and clinical sources in previous studies in Bangladesh17,18,21,22. Further, we found cross-source similarity in genetic sequences among the enteric bacteria. Fifth, resistance genes to various classes of antibiotics were commonly detected among isolates of E. coli, and V. cholerae, followed by Salmonella spp., and Shigella spp. Resistant phenotypes were commonly carrying genes to ceftriaxone (blaTEM, blaCTX-M-15, blaOXA), ciprofloxacin (qnrB), tetracycline (tetA) and cotrimoxazole (sul1) in the majority of the water sources. However, a resistant marker against colistin (mcr-1) was prevalent among samples from hospital wastewater, industrial effluent, municipality wastewater, and river water. Numerous studies have reported the presence of antimicrobial resistance genes (ARGs) to beta-lactam, sulfonamides, tetracyclines, chloramphenicol, aminoglycosides, and macrolides in water bodies worldwide. The majority of these studies considered the presence of ARG against synthetic antibiotics as a probable indicator of freshwater contamination11. The presence of both qnrA gene and mcr-1 gene in the isolates indicated probable contamination and mix-up of freshwater from different water bodies in nearby places.
We found both genotypic and phenotypic resistance patterns of E. coli and V. cholerae were closely related and distinct from Salmonella spp. and Shigella spp. in Bangladesh. These comparative findings are partially similar to previous studies in Bangladesh in clinical samples19,20,23,33. As this study lacked whole genome analysis, we focused on determining the association of specific genetic elements with phenotypic resistance in bacterial isolates. We found that the presence of randomly selected ARGs was associated with the phenotypic resistance of these bacteria, paralleling the findings of previous reports in Bangladesh and India19,20,23,30. Several studies have reported that horizontal gene transfer contributes to the dissemination of ARGs among different species of Enterobacteriaceae and freshwater is the hotspot of these ARGs transfer. In Australia and China, studies have found that MDR isolates of E. coli carry integrons capable of transferring resistance genes among different bacterial species11. Several studies have found that mobile genetic elements including, plasmid, transposons, and integrons contribute significantly to the horizontal transfer of these resistance genes. We also suspect the involvement of these mobile genetic elements in the transfer of ARG among different species of bacteria, which needs further justification in future studies.
Sixth, we found that bacterial isolates from hospital wastewater, industrial effluent, municipal wastewater, river water, and canal water displayed high levels of phenotypic resistance (40–60%) against most of the tested antibiotics, contrasting with previous studies28,29,30. Further, resistance against a broader antimicrobial spectrum, imipenem was found only among E. coli (100%) isolated from the hospital wastewater. Enteric bacteria from hospital wastewater, industrial effluent, and municipal sewage exhibited similar resistance patterns to co-trimoxazole, colistin, norfloxacin, and chloramphenicol. Our findings are in good agreement with the previous reports in India, China, and Thailand30,31,32. Cross-source differences in the prevalence of resistant bacteria were significant between water bodies getting antimicrobials directly and water bodies distantly situated from antibiotic exposure. In resource-limited settings, the implementation of ETP, and waste management systems are absent or weak. As a result, direct discharge of wastewater and effluent from hospitals, industries, animal farms, and municipality sewerage are getting huge amounts of antimicrobials and antimicrobial-resistant bacteria.
The swift and unchecked escalation in the application of antibiotics for human therapy, as well as in animal feed and treatments, has markedly influenced the proliferation of MDR, a finding backed by prior studies19,20,23,32,33. Multidrug-resistant isolates were most prevalent in hospital wastewater (55.84%) followed by municipal wastewater (44.29%), and industrial effluent (41.17%). This distribution is supported by previous studies in Ghana and Nigeria28,29. Contrarily, isolates from rivers, lakes, ponds, and irrigations exhibited no instances of multidrug resistance which is in consistent with previous studies in India28,29,30. Further, the mixing of contaminated water with rivers, canals, lakes, and ponds due to improper water management and distribution systems and during floods is contributing to increased concentration of ARBs in water. These findings will add new knowledge on AMR and MDR in water in central Bangladesh.
To the best of our knowledge, this study is one of the first studies in Bangladesh to comprehensively analyze antimicrobial resistance and multidrug resistance patterns in E. coli, V. cholerae, Salmonella spp., and Shigella spp. in environmental water sources. In the future, to get more comprehensive findings several factors should be considered. First, Minimum Inhibitory Concentration (MIC) testing can be conducted, which may provide a more detailed understanding of the bacterial isolates’ resistance levels. Second, the pathogenicity of the isolates can be determined. Third, an analysis of mobile genetic elements, such as plasmids or integrons involved in the dissemination of antimicrobial resistance properties to reveal the resistance mechanisms of the selected bacteria needs to be conducted. Fourth, a whole genome analysis of the isolates can be performed, which could add more accurate insight into the genotypic resistance and diversity of bacterial isolates. Fifth, a large sample should be studied to understand the diversity more precisely.
Findings from this work will enhance understanding of the antimicrobial resistance bacteria in water sources in resource-limited set-ups like Bangladesh. Additionally, an integrated method to assess the occurrence of antimicrobial resistance and multidrug resistance isolates has provided valuable insights into the domain. This investigation aids in partially identifying and comprehending the real situation of antimicrobial resistance and its molecular components spreading through water.
This study highlights the increasing level of antimicrobial and multidrug resistance in E. coli, V. cholerae, Salmonella spp., and Shigella spp. found in water sources, particularly in industrial and hospital wastewater in Bangladesh. These results identify these settings as key areas for the spread of antibiotic-resistant bacteria with genetic markers. Consequently, there is an imperative need for continuous surveillance of antimicrobial resistance in water bodies and the implementation of efficient waste management and monitoring measures to address this significant public health issue. Our findings highlight the improvement of strategies and infrastructure in industries producing and discharging antibiotics, hospitals, clinics, and places of antibiotic use and disposal into the environment. Integrated measures and policies for awareness building and intervention priorities should be martialized by involving policymakers, antibiotic producers, healthcare personnel, research scientists, and end-users to limit the dissemination of resistance in resource-limited developing and poor areas. The findings from this study establish that setting priorities and adopting strategies to limit the burden will require cross-source identification and proper assessment of the focal point of spread and implementation of multidisciplinary knowledge.
Methods
Ethical approval
This study did not involve any human or animal samples. We obtained the ethical clearance from the Biosafety, Biosecurity & Ethical Committee (BBEC) at Jahangirnagar University and the approval number for this study is BBEC, JU/M 2025/02 (202).
Method guidelines
The authors confirm that all of the used methods were performed in accordance with relevant and appropriate guidelines and regulations. All methods were performed following previously published articles from our laboratory19,20,33,34,35. For microbial culture and Kirby-Bauer disk diffusion, guidelines from ATCC (https://www.atcc.org/) and CLSI (https://clsi.org/) were followed and adopted, respectively.
Sampling site and sample collection
Fifty water samples were aseptically collected from 24 sites in Savar from January 2023 to December 2023. The study area was situated at coordinates 23.8583°N 90.2667°E, encompassing a residential count of 66,956 housing units within its expanse of 280.13 square kilometers. We collected 100 ml of water/per site and used an ice box for transporting the samples maintaining 4 °C temperature. The collection sites included industrial effluent, hospital wastewater, municipal wastewater, river water, canal water, lake water, pond water, and irrigation water. For different sources, we collected samples from three different sites. Further, from every sampling site we collected two samples. We used filtration, dilution, and direct drop plate inoculum according to the nature of the collected samples. A water sample of 1 ml was mixed with 9 ml of 0.85% sterile normal saline, followed by a serial dilution process ranging from 10−1 to 10−5. Subsequently, aliquots of 100 µl from the dilutions of 10−1, 10−3, and 10−5 were carefully spread onto sterile nutrient agar plates utilizing a sterile spreader. After a subsequent incubation at 37 °C lasting between 18 to 24 h, the individual colonies that emerged on each plate were meticulously tallied and the results were recorded. For filtration, we used 10 ml, 20 ml, 30 ml, 40 ml, and 50 ml water samples to pass through 0.45 µm cellulose paper and transfer the filter paper to agar plate media for growing the microorganisms.
One sample was collected during higher average atmospheric temperature (April-September) and another one during colder season (October-March). Collected samples were immediately transported to the laboratory following the appropriate transportation protocol. Upon arrival, physicochemical properties like temperature, pH, DO, and BOD were assessed before microbiological analysis commenced. All methods were performed following our previously published articles19,20,33,34,35.
Physicochemical and biological analysis of the samples
A mercury-in-glass thermometer ranging from 0 to 100 °C was utilized to ascertain the water temperature. Water pH levels were gauged using an ion-sensitive field-effect transistor pH meter (Orion-2 STAR, Thermo-Scientific, USA). The dissolved oxygen was measured in the water samples immediately upon collection with a DO meter (970 DO2 meter, Serial # 20600, Jenway, UK), ensuring no air was present by filling the sample containers to the brim. Biological Oxygen Demand (BOD) was determined by measuring the Dissolved Oxygen (DO) consumed by microorganisms in the sample over 5 days. The microbiological quality of the gathered water specimens was evaluated via previously established conventional microbiological techniques19,20,33,34,35.
Laboratory tests for enteric bacteria
Characterization of bacterial colonies was conducted on selective agar media designed for this purpose. MacConkey Agar, Thiosulphate-Citrate-Bile Salt Sucrose (TCBS) Agar, and Salmonella Shigella (SS) Agar media (HIMEDIA, India) were employed in a sterile manner for the isolation and identification of bacteria such as E. coli, V. cholerae, Salmonella spp., and Shigella spp. For further identification of E. coli, Eosin Methylene Blue (EMB) Agar (HIMEDIA, India) was used, while Xylose Lysine Deoxycholate (XLD) Agar (HIMEDIA, India) was applied for the identification of Salmonella spp. and Shigella spp19,20.
Antibiotic susceptibility test of bacterial isolates
The antibiotic susceptibility test was conducted using the Kirby-Bauer Disk Diffusion Test following the CLSI guidelines19,20,36. Eight distinct groups of antibiotics, as outlined in the CLSI manual, were employed in this testing (Supplementary Table ii). Bacterial susceptibility to each antibiotic was assessed by measuring the diameter of the inhibition zone, and results were categorized into resistant, intermediate, or susceptible based on CLSI criteria. The E. coli ATCC 25922 strain served as a control to verify the accuracy of the testing protocol19,20.
Bacterial genome extraction and polymerase chain reaction
The bacterial genome extraction was carried out by using the well-established boiled DNA method19,20,37. Bacterial isolation was achieved through polymerase chain reaction (PCR). For the PCR assays, sequences of 16S rRNA primers (F- AGT TTG ATC CTG GCT CAG and R- ACC TTG TTA CGA CTT) were utilized20. The composition of the PCR reaction mixture included 12.5 µl of 2X master mix (GoTaq Green Master Mix, Promega, USA), 1 µl each of the forward (F) and reverse (R) primers, 6.5 µl nuclease-free water, and 4 µl template DNA (Eppendorf, Germany), culminating in a 25 µl reaction volume. The PCR was executed in a thermal cycler (2720 Thermal Cycler, Applied Biosystems, USA), with the protocol setting at 94 °C for 5 min, then 35 cycles of 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 60 s8.
PCR reaction to detect antimicrobial resistance genes
For the blaTEM resistance gene (750 base pairs), primers F-TCGGGGAAATGTGCGCG and R-TGCTTAATCAGTGAGGACCC were used (Supplementary Table iii), with the PCR protocol including an initial 94 °C for 7 min, 30 cycles of 94 °C for 30 seconds, 53 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 5 min. Primer pairs F- GTTACAATGTGTGAGAAGCAG and R- CCGTTTCCGCTATTACAAAC were applied for the blaCTX-M-15 resistance genes, with conditions at 96 °C for 10 min, followed by 35 cycles at 94 °C for 1 min, 50 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 10 min (Supplementary Table iii). For amplifying the blaOXA resistance genes, primer pairs F- ACACAATACATATCAACTTCGC and R- AGTGTGTTTAGAATGGTGATC were utilized. The PCR procedure for blaOXA included an initial step at 96 °C for 5 min for one cycle, followed by 35 cycles of 96 °C for 1 min, 60 °C for 1 min, 72 °C for 2 min, and a final extension at 72 °C for 10 min. To amplify the qnrA resistance gene (492 base pairs), primer pairs F- GGATGCCAGTTTCGAGGA and R- TGCCAGGCACAGATCTTG were employed, with the thermal cycling conditions set at 94 °C for 5 min, then 35 cycles of 94 °C for 1 min, 59 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 10 min. The amplification of the qnrB resistance gene (469 base pairs) was achieved using primer pairs F-GATCGTGAAAGCCAGAAAGG and R-ACGATGCCTGGTAGTTGTCC (Supplementary Table 3), with PCR conditions of 94 °C for 2 min, followed by 35 cycles at 95 °C for 45 s, 53 °C for 45 s, 72 °C for 1 min, and a final extension at 72 °C for 5 min. For amplifying the sxt resistance gene (242 base pairs), primer pairs F- CATCTACCACTTCATAGGCAGC and R- CAGCTTAACTCACCAAGGAC were utilized. The PCR procedure for sul-1 included an initial step at 94 °C for 2 min for one cycle, followed by 35 cycles of 94 °C for 1 min, 60.5 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 10 min. Primer pairs F- GCTCGGTCAGTCCGTTTGTTCTTG and R- GGATGAATGCGGTGCGGTCTT were applied for the mcr-1 colistin resistance genes, with conditions at 93 °C for 3 min, followed by 35 cycles at 93 °C for 15 s, 57 °C for 30 s, 68 °C for 70 s, and a final extension at 72 °C for 5 min (Supplementary Table 3). All PCR products were stored at 4 °C19,20.
Agarose gel electrophoresis
The PCR amplicons were subjected to electrophoresis on a 1.5% agarose gel. This horizontal gel electrophoresis process was conducted for 30 min20. A DNA ladder of 1 kilobase pair was employed, and specific amplicons were made visible through the use of a UV spectrophotometer (SPECORD-205, Analytik-Jena, Germany).
Nucleotide sequence analysis
The nucleotide sequences of PCR amplicons (DNA) that tested positive for E. coli, V. cholerae, Salmonella spp., and Shigella spp. were sequenced using the Big-Dye terminator cycle sequencing kit along with an ABI Prism 310 Genetic Analyzer (Applied Biosystems Inc., Foster City, CA). These sequences underwent analysis through Chromas 2.6.5 (Technelysium, Helensvale, Australia). To confirm sequence homology, the BLASTn program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was utilized. Additionally, multiple sequence alignment (MSA) was executed using the ClustalW Multiple Alignment algorithm within the BioEdit 7.2.6 software38.
Phylogenetic analysis
The phylogenetic connections and molecular evolutionary studies of blaTEM, blaCTX-M-15, qnrA, and qnrB genes, compared with reference sequences, were performed utilizing MEGA 10.0 software39. Phylogenetic trees were constructed applying the Maximum Composite Likelihood (MCL) approach19. Reference sequences were obtained from the GenBank Database (https://www.ncbi.nlm.nih.gov/nucleotide/) (Supplementary Table 4). Data produced and/or examined in this study are accessible in the NCBI repository.
Determination of risk factors
For the source identification and MDR isolates dissemination, we integrated sets of variables that were potentially associated with circulation and selection for antimicrobial-resistant and multi-drug-resistant enteric bacteria in water sources (Supplementary Table 5). Different variables in this analysis were considered by following previous studies of our group and published guidelines on AMR in low- and middle-income countries by WHO3. For AMR, we first pooled enteric bacteria together and then applied antibiotic-specific models. For AMR, we included all eight sources in the model. In the later model of MDR, we only include four sources. Findings were documented in odds ratios (OR), with values of odds ratios >1.0 as a higher odd of the presence of antimicrobial-resistant/MDR-resistant isolates and <1.0 as a lower odd of the presence of resistance. Model fit was implemented by using McKelvey and Zavoina Pseudo R2, previously used and recommended for logistic multivariable models. The majority of values in McKelvey and Zavoina Pseudo R2 were ≥0.3 indicating good model fit40,41,42,43.
Statistical analysis
Categorical variables were represented as percentages, while continuous variables were described using mean/median values. The application of inferential statistics (p value) facilitated the analysis. The odds ratio (OR) for categorical variables was determined through two-tailed Chi-square or Fisher’s exact tests, accompanied by 95% confidence intervals (CIs). P values < 0.05 from two-tailed tests were deemed significant. Multivariable analysis was conducted to find out the association of source with the prevalence of isolates and antibiotic-resistant phenotype. Data analysis was conducted utilizing the Statistical Product and Service Solutions (SPSS v24.0) software (IBM, US).



Responses