Assessment of microbial communities affecting Almada Negreiros’ mural paintings at a Maritime Station in Lisbon

Introduction
The preservation of cultural heritage is a crucial endeavor in safeguarding both the tangible and intangible legacies of human civilization. Among these, mural paintings hold significant historical, artistic, and cultural value. Found in diverse contexts, including archaeological sites, religious buildings, and historical monuments, mural paintings are particularly susceptible to deterioration over time. While physical and chemical factors have been extensively studied as contributors to their degradation, the role of biological agents in biodeterioration has emerged as a critical area of research.
Recent advances in microbiology, molecular biology, and materials science have expanded our understanding of biodeterioration processes. As a result, biodeterioration is increasingly recognized as a critical factor in the overall deterioration of materials constituting cultural heritage artworks1,2,3,4. The proliferation of microbial communities on these surfaces can lead to both chemical5,6,7 and physical8,9,10 deterioration processes. Chemical deterioration primarily involves the release of harmful compounds resulting from microbial metabolism11,12,13,14. A typical example is the secretion of carotenoids, which oxidize and interact with pigments and binding agents, leading to discoloration7. Filamentous fungi and bacteria also produce extracellular enzymes and acids that break down complex organic compounds15 into simpler forms, which are then assimilated. These secretions can directly damage the substrate by weakening chemical bonds within the material or altering its physical properties16. Physical deterioration encompasses structural damage caused by microbial growth, such as the hyphae development of filamentous fungi17,18. When these fungi colonize a surface, their hyphae penetrate and physically disrupt the material matrix19,20. As they expand, they exert pressure on the surrounding substrate, leading to cracking, flaking, or detachment of surface layers21,22. In mural paintings, these processes can result in the loss of paint layers or mortar separation from the underlying support23. Additionally, microbial colonization can lead to biofilm formation, further intensifying deterioration by obscuring artistic details and diminishing the overall visual impact24,25.
Studies have also shown that microorganisms exhibit substrate specificity, meaning that the materials and pigments used in mural paintings influence the types of biological colonizers. Environmental monitoring is essential for murals that, due to their execution technique, history of moisture infiltration, structural characteristics, and geographical location, may be increasingly susceptible to this type of deterioration.
The focus of the current research is the mural paintings created by Almada Negreiros, one of the key figures of Portugal’s cultural and artistic scene of the 20th century. Two Portuguese maritime stations house significant mural painting by Almada Negreiros, created in 1945 and 1949. These stations contain a total of fourteen monumental murals, depicting Portuguese maritime culture, traditions, labour, representation of minorities and emigration. They are considered artistic landmarks of Portuguese modern art and the artist’s epitome, particularly the murals at Rocha do Conde de Óbidos26. Both sets of paintings adorn the west and east walls of the main departure lounge, located on the first floor of the buildings, which feature large windows. Over the years, they have been affected by water infiltration and have undergone at least two restoration campaigns (in 1971 and 1979)27,28. Along with three other murals painted by Almada in Lisbon between 1938 and 1956, these artworks have been the subject of study since 2021 within the project ALMADA-Unveiling the Mural painting Art of Almada Negreiros. Most of these murals exhibit signs of deterioration that require conservation intervention. Diagnostic surveys conducted at the Alcântara maritime station in 2021, and at Rocha in 2022 identified biocolonization in the paint layers26,29. The case study of the Maritime station of Rocha do Conde de Óbidos is particularly relevant for the extensive presence of organic material on the paint surface, which is currently being analyzed to determine its origin – whether from the original composition or past restoration interventions30.
Given the murals’ history of deterioration, including issues related to biocolonization, this study aims to thoroughly characterize the microbiome currently affecting them and assess its physical interactions with the painting materials. The station’s proximity to the Atlantic Ocean and the river exposes the murals to unique environmental conditions, such as salt spray and humidity fluctuations, which may further influence microbial growth. Additionally, the presence of organic material may unintentionally provide nutrients that promote this growth. By addressing these hypotheses, this study will contribute to future conservation efforts and aid in developing effective mitigation strategies.
Methods
Study site
The two maritime stations were already considered for previous studies27,28,29, and both – Alcântara and Rocha do Conde de Óbidos – are located along Lisbon’s riverside, near the Atlantic Ocean (Fig.1). Lisbon’s waterfront, stretching 19 km along the Tagus River, is directly exposed to environmental challenges that are exacerbated by climate change. Forecasts indicate a significant impact in the coming decades31,32, with risks including rising sea levels, coastal erosion, and increased heavy precipitation. These factors will accelerate the deterioration of concrete infrastructure and inevitably affect the mural paintings on the station walls.

a Portugal geographic location; b and c aerial view of Lisbon riverside and of the Maritime station of Rocha do Conde de Óbidos; d overview of the west wall of the main departure hall with mural paintings executed in 1949 by Almada Negreiros and measuring circa 7.80 ×3.80 m (images source: google earth and M. Ribeiro 2022).
According to the Portuguese Institute for the Ocean and Atmosphere (IPMA)33, Lisbon has a Mediterranean climate (Csa, Köppen classification), characterized by hot, dry summers and mild, wet winters. Summer temperatures typically range between 25 °C and 35 °C, while winters average between 10 °C and 15 °C, rarely dropping below 5 °C. The city’s proximity to the Atlantic Ocean moderates temperature fluctuations throughout the year. Annual rainfall averages around 700 mm, with precipitation concentrated in autumn and winter, mainly from November to February, while summer remain predominantly dry. The prevailing winds are northwesterly, especially in summer, bringing fresh air from the ocean. In winter, southwesterly and southerly winds can also occur, bringing humidity and storm activity. This climate is expected to change gradually in the coming decades, with projections indicating an increase in the number of hot days (maximum temperatures above 35 °C) and tropical nights (minimum temperatures above 20 °C), along with a substantial decrease in cold-weather events (e.g., frost days with minimum temperatures below 0 °C)31. Regional climate models also predict an increase in heavy daily precipitation events (above 10 mm/day) during winter31.
The two maritime stations housing Almada Negreiros’ mural paintings were built in the 1940’s and are among the earliest examples of reinforced concrete constructions in Portugal. As of 2022, both buildings exhibited structural damages, particularly at Rocha do Conde de Óbidos, where a lack of maintenance of expansive joints and rainwater drainage systems has contributed to deterioration34.
For the present study, the mural paintings were carefully examined, with particular attention to areas showing biodeterioration-related alterations such as color changes, flaking, and detachment of paint layers. On the west wall, which was the most affected by water infiltrations over the years, three murals – designated as P1, P2, and P3 – were analyzed, and six representative areas were identified for further investigation.
Sampling
Biocontaminants were collected from six representative areas exhibiting signs of contamination and alteration (Fig. 2a). Samples were obtained using non-invasive methods with sterile cotton swabs. To ensure representativeness, each sample was collected by swabbing three adjacent areas and was then transported in a sterile saline solution (0.9% NaCl) to the laboratory. Additionally, microfragments were gathered using fine tweezers to assess microbial-substrate interactions through scanning electron microscopy (SEM). The sampling process adhered to strict protocols designed to minimize both the structural and aesthetic impact on the artwork (Fig. 2b).

a Sampling points and b details of sampling at points 2, 4, and 5 respectively.
High-throughput sequencing (HTS)
A culture-independent method was adopted to obtain a broader view of the microbiota present on the mural paintings, including metabolically inactive microorganisms. Data analysis provided deeper insights into the predominant genera comprising both prokaryotic and eukaryotic colonizers.
DNA was extracted from the swabs previously placed in sterile saline solution (0.9% NaCl) and incubated in an orbital shaker at 100 rpm for 48 h. The extraction was performed using the DNeasy® PowerSoil® Pro Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The extracted DNA was quantified via spectrofluorimetry using the QuantusTM Fluorometer (E6150, Promega, Madison, WI, USA) and the QuantiFluorKit® One dsDNA System (Promega, Madison, WI, USA).
Following DNA extraction and quantification, amplification of the ITS2 region for eukaryotes and the 16S rDNA subunit (V3–V4 region) for prokaryotes was performed.
The amplification process was carried out using a Polymerase Chain Reaction (PCR) in a Thermocycler (MJ MiniTM Gradient Thermal Cycler, Bio-Rad Laboratories Inc., CA, USA). For bacteria amplification, the following primers were used: forward primer Bakt_341F (5′-CCTACGGGNGGCWGCAG-3′) and reverse primer Bakt_805R (5′-GACTACHVGGGTATCTAATCC-3′). For eukaryotic communities, a pool of forward primers was used: ITS3NGS1_F (5′-CATCGATGAAGAACGCAG-3′), ITS3NGS2_F (5′-CAACGATGAAGAACGCAG-3′), ITS3NGS3_F (5′-CACCGATGAAGAACGCAG-3′), ITS3NGS4_F (5′-CATCGATGAAGA ACGTAG-3′), ITS3NGS5_F (5′-CATCGATGAAGAACGTGG-3′), and ITS3NGS10_F (5′-CATCGATGAAGAACGCTG-3′) with the reverse primer ITS3NGS001_R (5′-TCCTSCGCTTATTGATATGC-3′).
For the DNA amplification of prokaryotic microorganisms, 5 µL of forward primer 341 F (1 mM) and 5 µL of reverse primer 805 R (1 mM), 10 µL of MyTaqTM HS Mix 2X (Bioline, Taunton, MA, USA) and 5 µL of extracted DNA were added to the PCR tubes, making a final volume of 25 µL. The PCR conditions included an initial denaturation at 95 °C for 3 min, followed by 25 cycles of 98 °C for 20 s, 55 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 5 min.
For eukaryotic microorganisms, the reaction mixture combined 6.5 µL of RNase-free water, 3.5 µL of the primer pool (10 nM), 10 µL of MyTaqTM HS Mix 2X (Bioline, Taunton, MA, USA), and 5 µL of DNA. The PCR program consisted of an initial denaturation at 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 sec, annealing at 55 °C for 30 sec, and polymerization at 72 °C for 20 sec, with a final extension at 72 °C for 2 min.
To confirm amplification and verify the integrity of the PCR products, 2% (w/v) agarose gel electrophoresis was performed using DNA Green Safe Premium dye (NZYtech, Lisbon, Portugal). Samples were loaded onto the gel alongside a 100 bp DNA marker, and electrophoresis was conducted at 90 V. After completion, the gel was revealed using a transilluminator (Bio-Rad Molecular Imager® Gel Doc™ XR+ Imaging System). Gel images are presented in the Supplementary Data (SD1 for Prokaryotic and SD2 for Eukaryotic).
The PCR products were purified using magnetic beads (High PrepTM PCR Clean up System Kit, MagBio Genomics, Gaithersburg, MD, USA) following the manufacturer’s instructions. The purified products were then quantified using the QuantiFluorKit® One dsDNA System (Promega, Madison, WI, USA) on the QuantusTM Fluorometer (E6150, Promega, Madison, WI, USA).
The amplified PCR fragments were indexed to encode the studied samples. The reaction mixture for this PCR contained 5 µL of each purified sample, 5 µL of each index (i7 and i5) (Nextera XT Index Kit, Illumina), 10 µL of RNase-free water, and 25 µL of MyTaqTM HS Mix 2X (Bioline, Taunton, MA, USA), making a total volume of 50 µL. Amplification was performed under the following conditions: 3 min at 95 °C, followed by 8 cycles of 30 s each at 95 °C, 55 °C, and 72 °C, with a final extension at 72 °C for 5 min. The products were then stored at 4 °C.
The libraries were normalized to a concentration of 4 nM, denatured, and diluted to a final concentration of 10 pM.
The sequencing process utilized the MiSeq Reagent Kit V2 (500) on the Illumina MiSeq system, with 20% PhiX control (Illumina). Sequencing was performed using a 2× 250 paired-end (PE) configuration, with image analysis and base calling carried out using MiSeq Control Software (MCS, version 4.1.0.656) directly on the Illumina MiSeq instrument (San Diego, CA, USA). The obtained sequences were demultiplexed using the CASAVA package (Illumina, San Diego, CA, USA), and reads shorter than 200 bases were discarded. Forward and reverse reads were merged using AdapterRemoval v2.1.5 software. The QIIME package v1.8.0 was used for Operational Taxonomic Units (OTU) generation, taxonomic identification, and the calculation of sample diversity and richness indices, following methodologies described in previous research5,35. Representative sequences of each OTU were selected for taxonomic assignment.
Scanning electron microscopy observations
Microbial-substrate interactions were investigated using a Field Emission Gun Scanning Electron Microscope (FEG-SEM) Tescan Clara (Tescan, Brno, Czech Republic). Microfragments were observed without prior treatment under the following conditions: 10 kV accelerating voltage, 100 pA beam current, 160 Pa chamber pressure, a working distance of 6–10 mm, and imaging with the backscattered electron (BSE) detector.
Results and discussion
Determining the microbiome inhabiting artworks provides essential insights for researchers and conservators, enabling the development of targeted conservation strategies. Since microorganisms can affect materials through chemical and physical processes, identifying the specific microbiome thriving allows conservation efforts to be tailored to mitigate their impact and prevent the loss of artistic details. While many microbial strains proliferating in a specific environment are viable and metabolically active, they may not be cultivable. Therefore, a culture-independent approach was employed in this study, taking advantage of the HTS method potentials.
HTS approach
Next Generation Sequencing (NGS) has proven to be a valuable technique for obtaining a detailed characterization of the microbiome, particularly on delicate materials such as artworks. The results indicate that Proteobacteria and Actinobacteria are the predominant phyla in the prokaryotic microbiome (Fig. 3), while Firmicutes, Cyanobacteria, and Bacteroidetes are also present in significant, though lesser, abundance. The dominance of Proteobacteria and Actinobacteria, known for their adaptability and resilience, suggests they may be primary contributors to biodeterioration. Many species within these phyla have been associated with the degradation of wall paintings and mortars36,37,38,39.
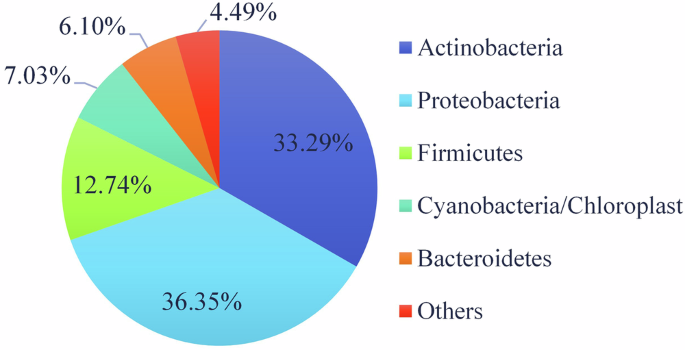
Predominant Phyla of the prokaryotic microbiome thriving on the mural paintings.
The diversity among bacterial groups suggests multiple degradation mechanisms, including acid production and biofilm formation, which can accelerate damage to murals. Figure 4 illustrates the distribution of prokaryotic genera, highlighting the dominant presence of Cellulomonas and Methylobacterium. Cellulomonas, recognized for its cellulolytic activity, likely contributes to the breakdown of organic binding materials, while Methylobacterium may influence biofilm formation and pigment alteration through its metabolic byproducts.
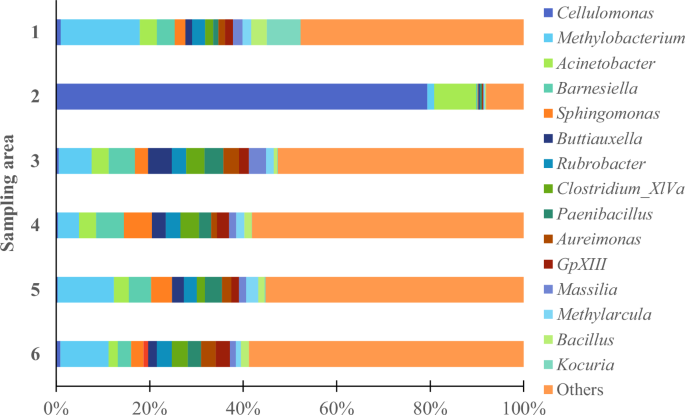
Relative abundance of the predominant bacteria (at genus level) thriving on the mural paintings.
In addition to these dominant genera, other bacterial groups – Acinetobacter, Barnesiella, Sphingomonas, Streptophyta, Buttiauxela, Rubrobacter, Clostridium XIVa, Paenibacillus, Aureimonas, GpXIII, Massilia, Methylarcula, Bacillus and Kocuria – were identified as prevalent colonizers in the selected areas. While their individual abundance is lower, their cumulative metabolic activities may significantly impact the integrity of the artworks.
The presence of cellulolytic bacteria, such as Cellulomonas, Clostridium, and Bacillus, is particularly noteworthy due to their ability to degrade cellulose-based materials, which are commonly found in mural painting substrates and organic binding media40. Additionally, Rubrobacter, Bacillus, and Kocuria have been strongly associated with discoloration and aesthetic alterations in both paintings and mortars41,42,43. These bacteria produce pigments and enzymes capable of modifying or degrading the original pigments in the artwork. For example, Rubrobacter, which thrives in extreme conditions, produces carotenoid pigments that can stain the artwork, while Kocuria may induce localized discoloration through its metabolic pathways.
Biofilm formation is another critical factor in the biodeterioration process. Genera such as Sphingomonas, Massilia, and Aureimonas secrete extracellular polymeric substances (EPS), facilitating microbial adhesion while trapping moisture and debris. These biofilms create microenvironments that promote microbial proliferation and accelerate chemical deterioration. The accumulation of biofilms also contributes to the peeling of pictorial layers and further destabilizes the substrate – a process exacerbated by the metabolic byproducts of genera like Acinetobacter and Paenibacillus. Furthermore, microbial activity influences mineralization and salt mobilization within the paintings. For instance, Methylarcula and Buttiauxela may interact with mineral components, potentially leading to efflorescence and structural weakening of the painted surfaces. These interactions highlight the multifaceted nature of biodeterioration, where both physical and chemical mechanisms contribute to deterioration. While Clostridium XIVa and GpXIII are less frequently mentioned in artwork biodeterioration, they may still play a significant role in these processes.
The results also indicate that the predominant phyla of the eukaryotic microbiome thriving on the mural paintings are Ascomycota (96.91%) and Basidiomycota (2.83%). Ascomycota has been widely reported as a dominant fungal group in mural paintings44, likely due to its wide distribution, airborne spore dispersal, and adaptability to nutrient-poor conditions, such as those found in these artworks.
Additionally, data analysis has provided further insights into the dominant fungal genera colonizing the murals (Fig. 5). The most relevant genera identified were Penicillium, Stachybotrys, Cladosporium, Aspergillus, Toninia, Malassezia, Memnoniella, and Verrucaria. Many species from these genera form filamentous or lichen-like structures, making them the most common deteriogens of wall paintings37,44,45.

Relative abundance of the predominant fungi (at genus level) thriving on the mural paintings. Samples 3 and 4 did not presented PCR amplification.
As previously mentioned, fungi exhibit strong enzymatic capabilities and have a high potential for biodeterioration4,46,47,48. Beyond inducing chemical deterioration, fungal growth can lead to mechanical impairments due to hyphal penetration into paint layers and the substrate. Over time, this activity can result in flaking, cracking, and powdering of the paint. Additionally, fungal biofilms can trap dust, dirt, and moisture49,50, creating an environment that fosters further microbial colonization, thus accelerating the deterioration process.
Among the fungal genera identified, Aspergillus, Penicillium, and Cladosporium are particularly associated with mural painting deterioration. Their resilience and adaptability enable them to thrive in diverse environmental conditions, where they contribute to deterioration by producing acids, spores, and pigments4,23,47,51,52,53. Furthermore, species from genera such as Stachybotrys, Memnoniella, Toninia, and Verrucaria were also found in significant abundance in specific areas. These fungi are known for their extracellular cellulase production, which facilitates the degradation of organic components54 in the paint layers. Notably, Stachybotrys has been associated with cellulose-based conservation treatments, as seen in a case study involving a subsurface medieval chapel, where residual cellulose in rock fissures provided a carbon source for microbial growth55. In this case, Stachybotrys was linked to a rosy discoloration, suggesting its involvement in pigment alterations.
Microbial-substrate interaction
The collected microfragments were examined to investigate the interaction between microbial colonizers and the substrate. The deleterious effects of fungal hyphae were unequivocally confirmed through SEM micrographs (Fig. 6). Given the inherently porous nature of mortars and paint layers, these materials provide an ideal environment for microbial proliferation. Consequently, the growth of filamentous fungi exerts mechanical pressure on the substrate, leading to the formation of microcracks in the paint layers and, subsequently, in the underlying mortar2. Over time, these microcracks can expand into larger fissures, ultimately resulting in flaking and detachment of pigment layers, a phenomenon widely reported in previous studies56,57,58. The extensive penetration of hyphae into deeper layers of the mortar exacerbates internal stress within the material, leading to structural weakening and eventual disintegration of the substrate. If no mitigation strategies are implemented, continuous microbial proliferation will likely contribute to the significant loss of the original artwork.

a and b Proliferation of conidiophores through the painting layers and (c) presence of fungi spores on painting layers’ surface.
Microfragments containing traces of organic material were also analyzed to gain deeper insights into microbial-substrate interactions in these artworks. The acquired micrographs established a clear correlation between organic matter and microbial colonization (Fig. 7). It is evident that microbial proliferation is preferentially localized on carbon-rich surfaces, as these serve as essential nutrient sources for microbial communities. Due to their diverse metabolic pathways, microorganisms can readily degrade organic molecules, extracting the necessary energy and building blocks for growth59. In the studied paintings, the primary source of carbon content detected on the paint surfaces was Gelvatol®, a polyvinyl alcohol (PVA) applied by spray during conservation and restoration treatments performed in the 1970’s to stabilize flaking and powdering paint layers29. However, the potential use of organic binders or coatings by Almada Negreiros cannot be ruled out. While the official documentation classifies the paintings at the maritime station of Rocha Conde de Óbidos as frescoes, recent studies suggest that the artist may have employed techniques beyond traditional fresco execution34. Fourier-Transform Infrared Spectroscopy (FTIR) scans conducted in 2023 revealed the presence of oil, protein, and oxalates in both deteriorated and well-preserved paint layers, alongside traces of PVA.

White narrows are signalizing those areas.
HTS identified the presence of Stachybotrys and Memnoniella species in the analyzed paint layers. Previous studies indicate that carbon-rich materials, such as cellulosic substrates, strongly stimulate Stachybotrys proliferation60, and most Stachybotrys species are known to colonize cellulose-rich environments61. Notably, Stachybotrys and Memnoniella are considered closely related due to their morphological, biological and phylogenetic similarities. Could the presence of these fungi be linked to traces of cotton, possibly used in past cleaning interventions on the painted surfaces? So far, no historical documentation has been found to confirm this hypothesis.
Additionally, the lichen-forming fungi Toninia, and Verrucaria62 were identified among the eukaryotic colonizers. Lichenized fungi play a key role in microbial communities due to their mutualistic association with photobionts63. This symbiosis allows lichens to provide structural support and protection to microbial proliferation, while also breaking down minerals and organic matter, releasing essential nutrients that can be utilized by other microorganisms. Given these capabilities, lichen-forming fungi likely contribute to microbiome dynamics within the studied artworks and may influence biodeterioration processes.
These findings underscore the urgent need for conservation actions to prevent the permanent loss of these invaluable artworks. Traditional conservation treatments, historically challenging to conservator-restorers, often rely on chemical agents, which can be harmful to both the environment and practitioners. Additionally, some treatments may pose risks to the artworks itself, potentially leading to further deterioration over time. As a result, there is a growing shift toward environmentally friendly conservation approaches64, which prioritize sustainability while ensuring effective preservation. These innovative methods aim to mitigate microbial deterioration using less invasive and less harmful compounds, promoting safer and more long-term conservation strategies. Their exploration and implementation should be actively encouraged to ensure the preservation of cultural heritage.
In resume, this study provides valuable insights into the mechanisms of deterioration, with a particular focus on biodeterioration. NGS analyses identified the predominant microbiome present on these artworks. Among prokaryotes, the dominant phyla included Proteobacteria and Actinobacteria, while Ascomycota was the most prevalent eukaryotic phylum. Several key genera were detected, including Cellulomonas, Methylobacterium, Rubrobacter, Penicillium, Cladosporium, Aspergillus, and Stachybotrys all recognized for their biodeterioration potential. SEM micrographs provided visual evidence of filamentous fungi inflicting mechanical damage to the paintings. The extensive hyphal growth contributed to the formation of cracks, flaking, and detachment of the pigment layers. The study further revealed that carbon-rich organic materials, whether from the artist’s original application or previous conservation treatments, played a crucial role in fostering microbial growth. This is supported by both SEM imaging, which highlights microbial colonization on organic-rich areas, and sequencing data, which identified organic matter-decomposing microorganisms, even at lower abundances. Given these findings, the implementation of carefully selected mitigation strategies is strongly recommended. Effective control of biodeteriogenic agents is essential to preserve the integrity of these monumental mural paintings, which are housed in a historically significant building in contemporary Portuguese history.





Responses