B(RAIN)2—BRAIN integrated Resource for Anatomy and Intracranial Neurophysiology

Background & Summary
Human intracranial electroencephalography (iEEG) recordings, combined with neuroimaging data, are pivotal to enhancing our understanding of the human brain and addressing the pathophysiology and treatment of disorders such as Parkinson’s disease (PD), epilepsy, and depression.
Because such data can only be acquired in patients undergoing neurosurgical procedures, and due to the specialized effort, and inherent risk associated involved in collecting such data, iEEG recordings are relatively rare and require years to acquire in meaningfully sized cohorts of patients, increasing the importance and value of sharing such data. Sharing these data, however, is particularly challenging because iEEG datasets vary widely in structure, with different centers using unique organization methods, protocols, tasks, and conditions, complicating collaboration and reproducibility. Existing file formats often miss key details needed for analysis, such as electrode positions, referencing methods, and physiological data, highlighting the need for a standardized approach to fully describe iEEG data and its context. In response to these challenges, the BRAIN integrated Resource for human Anatomy and Intracranial Neurophysiology (B(RAIN)2) was developed as a multi-center data resource, providing standardized imaging and intracranial neurophysiological data from patients who have undergone deep brain stimulation (DBS) implantation with iEEG recordings.
The BRAIN 2025 report emphasizes the critical need to maximize the scientific opportunities offered by neurotechnology developments, particularly when working with human intracranial data1. This aligns with the broader goals of the National Institutes of Health (NIH) Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Initiative, which has made significant investments in intracranial neurophysiological investigations to advance our understanding of motor control, memory, language, and cognition2,3,4,5. Despite the growth in such studies, human intracranial neurophysiological data remains relatively rare and extremely valuable. The NIH has responded to this by investing in the public archiving of these data through the Data Archive for the BRAIN Initiative (DABI)6. However, the data’s utility for large-scale studies remains limited by the lack of standardization and integration across different sites.
A key challenge in advancing our understanding of neural circuits is the significant variability in neural oscillation data across subjects, with unclear sources of this variability. For instance, the beta peak varies widely among individuals, yet the underlying causes remain largely unexplored7,8,9. Neuroanatomy, spatial localization, and morphometric characteristics likely contribute to neural oscillations emerging from local and remote neuronal populations10,11,12. Although age-related and disease-related changes in brain oscillations have been studied13,14,15,16,17, the interaction between morphometry, disease, age, and neural signals is poorly understood.
Most studies to date have relied on electroencephalography (EEG) and magnetoencephalography (MEG), which, despite their non-invasive nature, suffer from poor localization and spatial specificity, particularly for signals arising from medial brain structures18,19. These limitations hinder the accurate investigation of anatomy-function interactions, especially compared to the precision offered by intracranial recordings. Additionally, much of the existing literature focuses on healthy subjects, with few studies examining the critical interactions between disease, morphometry, and neural oscillations20.
To overcome the challenges of standardization and integration, extensions to the Brain Imaging Data Structure (BIDS) were developed. Initially proposed for magnetic resonance imaging (MRI) data, BIDS has become a community standard for organizing and sharing brain imaging study data across common imaging modalities21. Its success in managing heterogeneity, ensuring straightforwardness, and reducing potential data-handling mistakes led to the creation of extensions for electrophysiological data, including MEG, EEG, and iEEG22,23,24. The scientific community was initially introduced to MEG-BIDS, which laid the groundwork for subsequent extensions. Building on this foundation, EEG-BIDS was developed to encompass EEG data management, and it was introduced alongside the iEEG-BIDS extension.
While these extensions have advanced data standardization, large, integrated datasets are critical for further advancing this field, but collecting sufficient data from a single institution is challenging. Platforms such as the Data Archive for the BRAIN Initiative (DABI), Distributed Archives for Neurophysiology Data Integration (DANDI), OpenNeuro, and Brain-CODE have overcome this by enabling collaboration across institutions while providing robust infrastructures for organizing and sharing neurophysiological and neuroimaging data6,25,26,27 For instance, DABI focuses on intracranial neurophysiology, supporting multiple data formats and strongly encouraging the use of Neurodata Without Borders (NWB) and BIDS standards21,28. Similarly, DANDI offers a repository for human and non-human neurophysiology and neuroimaging data, promoting BIDS and NWB. OpenNeuro, one of the largest neuroimaging repositories, improves accessibility by supporting iEEG data through the iEEG-BIDS extension. Meanwhile, Brain-CODE integrates diverse data types, including MRI and clinical measures, to secure data sharing and collaboration29.
Given the significance of iEEG data, and the challenges associated with data sharing, and integration, we developed B(RAIN)2 to directly address these issues by providing a comprehensive internal pipeline and database in an integrated BIDS version (https://dabi.loni.usc.edu/brain2). Building on existing efforts, B(RAIN)2 integrates anatomical and intracranial neurophysiological data from multiple sites, offering preprocessed and standardized neuroimaging and electrophysiological data for advanced brain structure and function exploration. This resource includes electrophysiological data organized using the iEEG-BIDS extension alongside a complete imaging BIDS directory for each subject. The directory encompasses high-quality multimodal neuroimaging data captured across preoperative, intraoperative, and postoperative stages, paired with electrophysiological recordings from both cortical and subcortical regions. All data are systematically structured within the BIDS framework to ensure consistent organization and accessibility (Fig. 1). This comprehensive resource enables deeper investigations into the interactions between anatomy, disease, and neural activity, offering a unique and powerful tool for advancing our understanding of the brain.
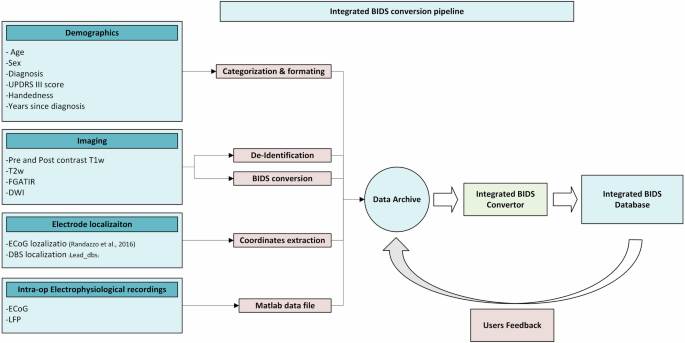
Overview of B(RAIN)2 Workflow for Integrated BIDS Directory Production.
Methods
Patient characteristics and cohort
For B(RAIN)2, we primarily focus on subjects within DABI who meet the inclusion criteria for the database6. These subjects must have available demographic information, oscillatory data from either or both cortical and subcortical structures, and sufficient neuroimaging data for electrode localization. No additional exclusion criteria were applied beyond the specified inclusion criteria. Randomization and blinding were not applicable, as this study is a retrospective analysis of existing clinical data.
The first B(RAIN)2 dataset release includes 45 patients30, and the second dataset release adds 30 more31, bringing the total to 75 patients with neuroimaging and iEEG recordings in integrated BIDS format, all diagnosed with Parkinson’s disease (PD) or essential tremor (ET). The next two releases are expected to include a total of approximately 67 additional patients. Beyond these, further data releases are anticipated, with more subjects continuously integrated into the database as part of ongoing collaboration efforts with DABI. B(RAIN)2 leverages DABI’s existing archive, which holds data from 1640 human subjects across 48 institutions, 987 of whom have high-resolution imaging or localized electrodes. B(RAIN)2 focuses on converting, preprocessing, and releasing data from this archive that meets its criteria. Additionally, the initiative is committed to enhancing its database with anonymized data from non-BRAIN-funded intracranial studies conducted at other centers, including but not limited to UT Southwestern Medical Center.
Each data provider contributing to DABI, and subsequently to B(RAIN)2 via DABI, has obtained patient consent through their respective Institutional Review Boards (IRBs).Participants consented both to take part in the research and to have their anonymized data openly shared. Those included in the current datasets from B(RAIN)2 have an age range of 33–81 years. For any future contributions to the database that include participants under the age of 18, parental consent will be required and documented by the respective contributing sites. The data owners have the responsibility to remove protected health information (PHI) before data sharing. B(RAIN)2 enforces strict deidentification, defacing, and anonymization protocols to guarantee that all data is fully deidentified before being publicly available.
Data preprocessing
The methodology for the B(RAIN)2 database is structured to comprehensively explain the preprocessing and organization of neuroimaging and electrophysiological data. This process is supported by an internal pipeline developed specifically for B(RAIN)2 to ensure the standardization, quality control, and organization of all data before sharing with the research community. Given the complexity and variability of iEEG and neuroimaging data, the pipeline is designed to address specific requirements for diverse datasets and acquisition modalities. While highly effective for current datasets, the pipeline is tailored to internal workflows and is not yet a generalized tool for external use. Future development aims to refine and validate the pipeline to accommodate broader data types and enable independent implementation across different research groups.
To accommodate the range of data collected, the workflow is divided into two key sections:
A. Neuroimaging Preprocessing: Manages the conversion, preprocessing, and integration of multiple imaging modalities
B. Electrophysiological Preprocessing: Focuses on the conversion and organization of iEEG recordings.
This dual approach ensures that imaging and electrophysiological data are standardized, anonymized, and prepared for in-depth analysis while fully complying with the BIDS guidelines.
Neuroimaging preprocessing
The neuroimaging data preprocessed in the B(RAIN)2 pipeline include preoperative MRI scans such as T1-weighted (T1W), T2-weighted (T2W), Fast Gray Matter Acquisition T1 Inversion Recovery (FGATIR), and Diffusion Weighted Imaging (DWI). Intraoperative imaging for lead localization is provided either by O-arm CT or fluoroscopy, depending on the centre’s protocol. The pipeline also incorporates pre- and postoperative axial CT scans with 1 mm slices for detailed bone and tissue density. Given the multi-center nature of the data, imaging sequences may vary across datasets, and certain modalities may be absent for some patients based on the protocols of the contributing centers.
Neuroimaging data organization and preprocessing are structured into three stages:
-
1.
Preprocessing and conversion to BIDS
-
2.
Electrocorticography (ECoG) localization
-
3.
DBS lead and Stereo-Electroencephalography (sEEG) Localization
All imaging data are anonymized during DICOM to NIfTI conversion, with further privacy protection using pydeface to remove facial features from MRI scans32.
Preprocessing and conversion to BIDS
A systematic approach is essential for converting MRI data from the Digital Imaging and Communications in Medicine (DICOM) format to BIDS (Fig. 2)33, ensuring standardized organization. Initially, DICOM data are inspected using tools such as Onis, Horos, or Osirix to verify imaging integrity34,35. DICOM files are converted to the Neuroimaging Informatics Technology Initiative (NIfTI) format, with relevant JavaScript Object Notation (JSON) files generated and data anonymized using MRIcron36. Facial features are removed from the MRI scans to enhance privacy protection. The imaging data are then renamed and structured according to BIDS guidelines using custom scripts. Compliance with the BIDS standard is verified through the BIDS-validator, ensuring accurate and standardized conversion of MRI data to the BIDS format37.
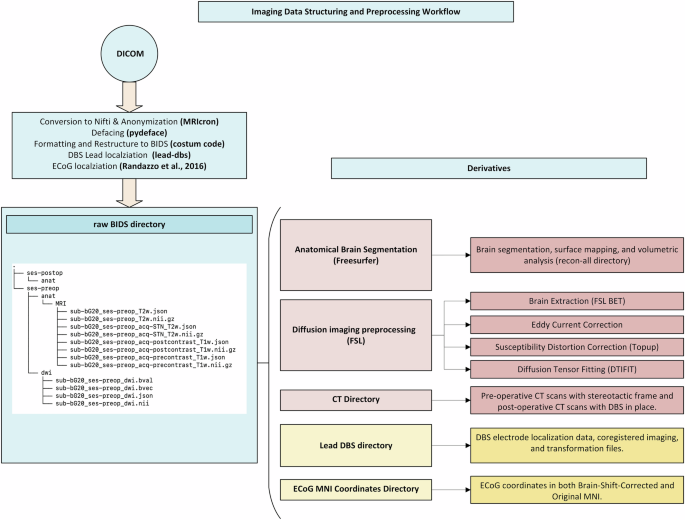
Overview of imaging data conversion, preprocessing, and BIDS structuring workflow.
DWI Data preprocessing
DWI preprocessing is performed using the FMRIB Software Library (FSL) version 6.0.6.438. Key steps include brain extraction using BET, correction of eddy currents and motion with the eddy tool and fitting the diffusion tensor model using dtifit to generate Fractional Anisotropy (FA) and Mean Diffusivity (MD) maps.
Anatomical data preprocessing
High-resolution T1-weighted MRI data undergo cortical reconstruction and volumetric segmentation using FreeSurfer version 7.3.2 (Martinos Center for Biomedical Imaging, 2023)39. The recon-all command facilitates motion correction, Talairach transformation, brain stripping, cortical reconstruction, and subcortical segmentation, providing detailed anatomical measurements for each subject.
Electrocorticography (ECoG) localization
ECoG localization is performed using a novel semi-automatic interface40, integrating intraoperative fluoroscopy or O-arm imaging with routine clinical scans, including preoperative MRI and pre-/postoperative CT images. CT scans are coregistered with the anatomical MRI using the Statistical Parameter Mapping (SPM) package (SPM12, http://www.fil.ion.ucl.ac.uk/) normalized mutual information method. A custom MATLAB interface visualizes and marks electrode locations on the coregistered images.
For fluoroscopy-based localization, the 2D fluoroscopic image is aligned with the 3D cortical and skull surfaces reconstructed in 3D Slicer41. The O-arm imaging method builds upon this approach, enhancing precision by providing a more precise 3D visualization of the ECoG strip in situ, further refining electrode localization. The final coordinates of each electrode are reported in the subject’s native space (Fig. 3).
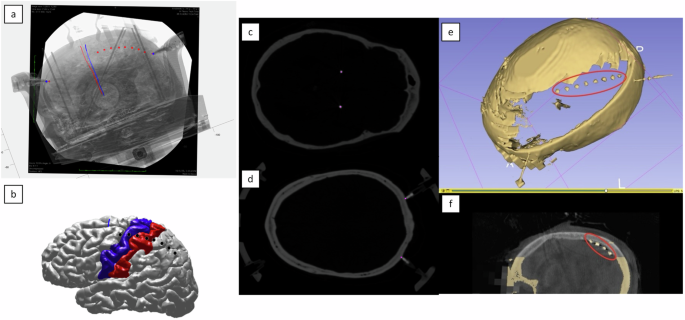
Steps involved in ECoG localization. (a) The final alignment of DBS electrode paths and stereotactic frame pin tips on the intraoperative fluoroscopic image based on coregistered CT scans, with manual pinning of ECoG locations on the fluoroscopic image. (b) Visualization of ECoG electrode locations projected onto a reconstructed cortical surface, segmented using the FreeSurfer Desikan-Killiany-Tourville (DKT) atlas. (c,d) Localization marks for DBS electrode tracts on postoperative CT and stereotactic frame pins on preoperative CT, respectively, with violet dots indicating electrode and pin locations. (e,f) Skull removal near the in situ ECoG strip and noise reduction, producing a unified 3D model of the ECoG strip and skull for final localization within the same anatomical space for cases using O-arm imaging instead of intraoperative fluoroscopy.
ECoG electrodes, initially measured in Right-Anterior-Superior (RAS) space, are transformed into the standard Montreal Neurological Institute (MNI) space using the Advanced Normalization Tools (ANTs) non-linear Symmetric Normalization (SyN) registration algorithm42. Each subject’s T1 image is registered to the MNI template, and the computed transformations are applied to the ECoG coordinates. To correct for brain shift, electrode locations are projected onto the brain surface following Hermes et al.43. Native ECoG coordinates are stored in the raw directory, while transformed, and brain-shift corrected data are stored in the derivatives directory, ensuring comprehensive data accessibility (Fig. 4). While this figure illustrates the transformation process for a single subject, electrode locations for all subjects are systematically stored in the database and are accessible in both native and MNI spaces for further analysis.
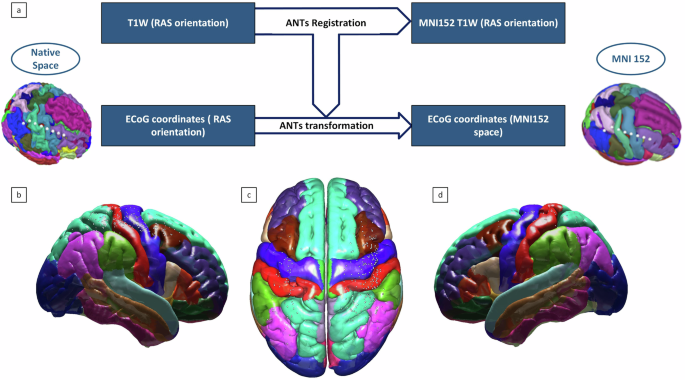
Visualization of ECoG electrodes mapped to MNI152 space for the B(RAIN)2 Dataset 1. (a) Schematic representation of the ANTs non-linear transformation workflow, mapping subject-specific electrode locations from native RAS space to MNI152 space. (b–d) Group-level visualization of ECoG electrodes across the cortical surface in MNI152 space, reflecting the overall distribution and coverage from the dataset. Electrode coordinates for all subjects in both native and MNI spaces are available in the shared database.
DBS lead and stereo-electroencephalography (sEEG) localization
DBS and sEEG localization are performed using the Lead-DBS toolbox44, which reconstructs the electrode trajectory and contact locations based on postoperative CT images. T1w, T2w, FGATIR, and 1 mm axial postoperative CT images are coregistered, corrected for brain shift, and pre-processed within Lead-DBS using ANTs Normalization. Final electrode positions are manually aligned and contact point coordinates are extracted in both native and MNI space and systematically stored in the shared database (Fig. 5). While the current dataset includes DBS only, sEEG data are not present. The described methodology is designed to accommodate both DBS and sEEG localization should such data become available in future datasets.
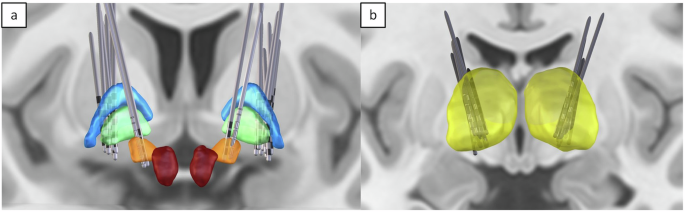
Group-level visualization of DBS electrode locations in the B(RAIN)2 Dataset 1, reconstructed in MNI space using Lead-DBS. (a) Globus pallidus internus (GPi) and subthalamic nucleus (STN) DBS lead placements in PD cases. (b) Thalamic DBS lead placements in ET cases. This figure illustrates the spatial distribution of DBS leads across subjects, highlighting anatomical targets for different conditions. Electrode coordinates for all subjects are systematically organized in the dataset in both native and MNI spaces.
Electrophysiology preprocessing
Converting raw electrophysiological recordings to the iEEG-BIDS format eases data curation and organization, especially for extensive ECoG and DBS data. This standard streamlines the data integration process across different modalities and studies, simplifying analysis and enhancing the investigative potential of the collected recordings22.
The conversion process within our pipeline encompasses a series of steps designed to transition raw electrophysiological recordings into a structured iEEG-BIDS format. Initially, a comprehensive review of the existing metadata of raw files is conducted to create a list of ‘required’ and ‘recommended’ fields in various components of iEEG-BIDS metadata, such as the sidecar JSON file, Channel description, and Electrode description, among others. Subsequently, reference documents for metadata common across all patients are generated to ensure a standardized approach to data handling.
Initially, recorded raw data is decomposed into essential functional modules to improve data organization. This segregation addresses the format diversity in acquisition software, such as BCI2000, AlphaOmega, or other acquisition software, which may vary depending on the recording utilities, tasks, and conditions.
This process employs a series of MATLAB classes that create data objects from raw data (both.mat and.dat files). These objects are then converted into BIDS-compliant Brain Vision Core Data format, where each recording comprises a Header File (.vhdr), a Marker File (.vmrk), and a Data file (.eeg). The intermediary events and data objects enable easy addition of future data formats or output models, ensuring data compatibility and accessibility for future analytical endeavours.
Specific segments, like the ‘Rest’ portion of behavioural tasks, are seamlessly extracted from principal datasets, enabling the swift creation of customized datasets for distinct secondary projects. Additional branches of the pipeline collect the experimental and organizational information to populate all the metadata BIDS file structure, including the automated extraction of organization information, task properties, and ECoG and DBS electrode details.
Lastly, all required fields for the sidecar JSON file, Channels description, Electrode description, Coordinate system JSON files, and event files are populated in compliance with the iEEG-BIDS format, ensuring a well-organized, standardized, and BIDS-compliant dataset ready for further analysis and sharing within the scientific community (Fig. 6).
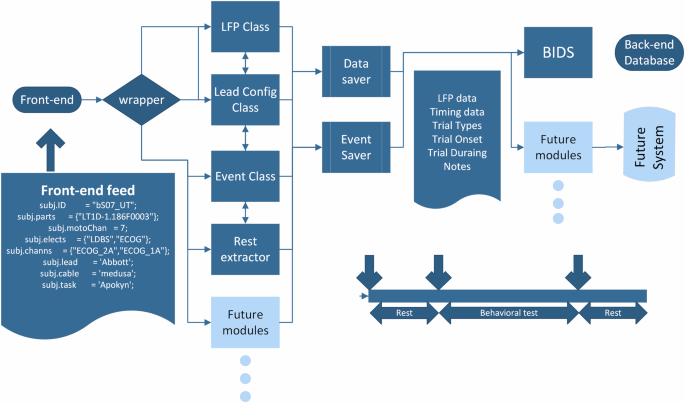
Illustrative Pipeline Showcasing the Conversion of Behavioural and Raw Electrophysiological Recordings to iEEG-BIDS Format.
Data Records
The data records associated with this study correspond to the cohort described in the Patient Characteristics and Cohort section, including subjects with iEEG recordings from B(RAIN)2 30,31, which leverages DABI’s existing archive and data from non-BRAIN-funded intracranial studies conducted at other centers. The datasets are stored in an integrated BIDS directory structure, ensuring standardized organization and accessibility6. Key data types include Neuroimaging Informatics Technology Initiative (NIfTI) files for neuroimaging and iEEG files in EEG, Vision Header File (VHDR), and (Vision Marker File) VMRK formats, along with associated metadata in JSON and Tab-Separated Values (TSV) formats (Table 1). All data formatting and structure are based on the BIDS specification for imaging and the iEEG-BIDS extension21,22. The B(RAIN)2 database and associated resources, including protocols and comprehensive data, are publicly available through DABI at https://dabi.loni.usc.edu/brain2, hosted on DABI’s website.
Technical Validation
All imaging data were manually inspected at multiple stages: pre-conversion, post-conversion, and post-defacing, ensuring the integrity and quality of the images throughout the process. Additionally, the FA maps and preprocessed DWI images were manually reviewed to confirm that standard quality was preserved.
For electrophysiological data, both ECoG and DBS electrode locations were carefully validated. ECoG locations were inspected manually at two critical stages: post-localization in native space and post-transformation into MNI space with brainshift correction, using a custom-developed code. DBS electrodes were evaluated using the built-in 2D and 3D viewers within the Lead-DBS toolbox following localization.
A custom MATLAB-based code using signal processing and Fieldtrip toolbox45 was developed for the quality control and visualization of electrophysiological data, including both ECoG and DBS recordings. The tool allows for the visualization of raw data across all channels with optional high/low pass filtering, enabling a comprehensive inspection of the signal quality. Additionally, the tool performs frequency-domain analysis using a multi-taper method to generate power spectral density (PSD) plots. These capabilities facilitate thoroughly validating data integrity before proceeding to in-depth analyses. (Fig. 7) illustrates the output of this tool, showcasing both the raw data visualization and the frequency spectrum analysis.
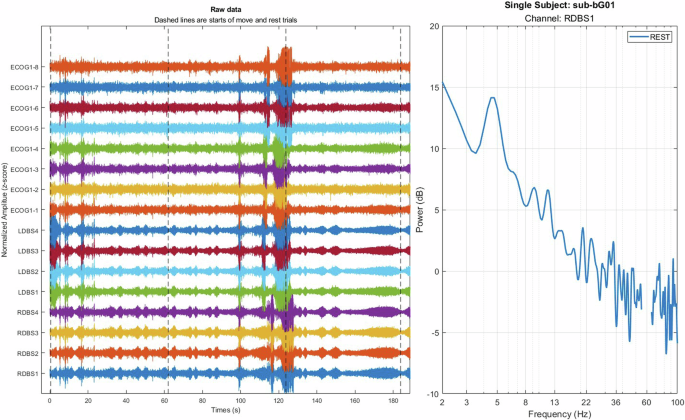
Visualization and frequency analysis of electrophysiological data. (Left) Raw ECoG and DBS signals across multiple channels are normalized for visualization. Dashed lines indicate the start of the trials. (Right) Power spectral density (PSD) plot for the RDBS1 channel during a rest trial, displaying power distribution across frequencies from 2 to 100 Hz.
Each dataset has been and will be tested with a BIDS validator to ensure structural and functional validity according to BIDS specifications37. For reproducibility, the BIDS reader and visualizer are available at: https://github.com/B-RAIN-2/BRAIN-2-Quality-Control-and-Analysis-Toolbox.
Usage Notes
The B(RAIN)2 will be freely available through DABI website. Researchers can create a free account on the DABI platform and request access by signing a Data Use Agreement (DUA) and briefly describing their project or study. Upon approval, access will be granted. All datasets are fully deidentified and provided strictly for scientific, teaching, or clinical research purposes, with safeguards ensuring data security and privacy. We hope these resources will foster innovation in developing novel tools for advancing clinical research and education.
As B(RAIN)2 is a multi-center database, certain limitations arise due to center variability.
Firstly, not all centers follow the same imaging protocols. While all subjects have high-resolution anatomical imaging for localization and anatomical studies, some may lack common modalities such as T2W, FGATIR, or DWI scans. Additionally, image resolution may vary depending on the year of acquisition or the specific center. Despite the strict inclusion criteria outlined in the patient characteristics and cohort sections, some centers may not provide complete demographic information due to variations in data sharing practices or acquisition protocols.
Secondly, although BIDS covers many imaging modalities and is a widely adopted formatting standard, it does not support certain imaging types, such as CT scans, or specific modalities like FGATIR. In these instances, we followed the BIDS naming conventions to maintain consistency, even though these modalities are not officially supported and did not fully align with BIDS validation criteria. We chose not to exclude these data, as they remain essential for completeness and research purposes.



Responses