Characterization of a SARS-CoV-2 Omicron BA.5 direct-contact transmission model in hamsters

Introduction
Since its emergence in 2019, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) continues to circulate and cause coronavirus disease-2019 (COVID-19)1,2. Novel variants with point mutations, insertions, and/or deletions in the viral genome, resulting in antigenic changes in the spike protein (S), have evolved since then. Infections with the Delta variant led to higher mortality and more intensive care unit (ICU) admissions compared to earlier variants3,4. The first antigenically distinct Omicron subvariant (BA.1) appeared at the end of 2021 and rapidly replaced Delta as the dominant variant worldwide by causing frequent breakthrough infections. This variant harbored 35 amino acid mutations in its S protein compared to the ancestral virus, which led to significant evasion of neutralizing antibodies elicited by prior vaccination or infection, and increased transmissibility in partially immune populations5,6,7. Many Omicron subvariants, such as BA.1, BA.5, and the currently circulating JN.1 sub-lineage KP.3, have since been identified and replaced prior circulating strains8. Disease severity and mortality associated with the Omicron variants were lower as compared to earlier variants. It is difficult to discern whether this observation is associated with changes in viral virulence, with increased population immunity due to vaccinations and natural infections, or a combination of the two. Although vaccines have been effective in protecting from severe COVID-19, the continuous evolution of SARS-CoV-2 requires yearly vaccine updates. Several in vitro and in vivo studies have demonstrated that therapeutic monoclonal antibodies are less effective against sub-lineages of the Omicron variant9,10,11. A supply of potent therapeutics that are resistant to antigenic drift is needed12.
To study intervention strategies against SARS-CoV-2 and its variants, well-characterized animal models are indispensable. Multiple species, including non-human primates, ferrets, hamsters, and (transgenic) mice, have been used13,14,15,16. Whereas the susceptibility of mice to SARS-CoV-2 is variable and depends on the genetic background and viral variant, ferrets and hamsters are naturally susceptible to most variants17,18. Infected ferrets shed SARS-CoV-2 from the upper respiratory tract but develop limited clinical signs, making it a suitable model for asymptomatic or mild disease in humans14. Golden Syrian hamsters more often develop severe disease after infection, partly due to similarities in tissue distribution of the entry receptor ACE2 between hamsters and humans19,20. Upon experimental infection, hamsters often exhibit high viral loads in the upper and lower respiratory tract, alongside clinical manifestations such as transient weight loss, respiratory distress, and histopathological changes in the respiratory tract. These included pulmonary consolidation, edema, and necrosis. Taken together, hamsters are the most used small animal model for studying SARS-CoV-2 and novel intervention strategies.
SARS-CoV-2 can be transmitted through direct or indirect contact with infected individuals. Viral particles are spread via respiratory droplets and aerosols during regular breathing, coughing, or sneezing21. Previous studies have shown that when modeling transmission in animals, directly inoculated donor hamsters efficiently transmit variants of SARS-CoV-2, such as the Beta or Delta variant, to naïve hamsters by direct contact or via the air3,22. Compared to an experimental SARS-CoV-2 inoculation into the nose or throat of hamsters, exposure of recipient animals to infected donors closely mimics the infection dynamics in humans, making transmission models more relevant for intervention studies. Body weight, viral loads and inflammation in alveoli but not bronchi, and infectious viral loads in the lung and nasal turbinates are significantly lower in Omicron BA.5-infected hamsters compared to earlier variants such as Delta23. Transmission potential in hamsters of Omicron BA.1 and BA.2 is lower compared to D614G, and lower viral loads in Omicron BA.5-infected hamsters might decrease transmission efficacy similarly24. This highlights the need for the establishment of well-characterized transmission models for individual SARS-CoV-2 variants.
Here, we established a SARS-CoV-2 Omicron BA.5 direct-contact transmission model in hamsters that can be used to evaluate interventions. First, we compared two intranasal doses of SARS-CoV-2 D614G and Omicron BA.5 in a small volume inoculum (10 µl per nostril) in male and female hamsters, to determine a dose sufficient for productive infection. Next, we determined the optimal co-housing time and duration of directly inoculated donor hamsters with immunologically naïve recipient hamsters to ensure consistent transmission. Finally, we compared viral kinetics and histopathology in tissues of the respiratory tract of donor and recipient hamsters.
Materials and methods
Animals
5–6-week-old female and male Syrian golden hamsters (Mesocricetus auratus) were obtained from a commercial breeder (Janvier labs, France). Animals were housed in groups of 3 in filtertop cages (425 × 266 × 185 mm; eurostandard type III, tecniplast), inside negatively pressurized HEPA-filtered isolators and had an acclimatization period of at least 7 days. During cohousing, animals were housed in groups of 4. Access to food and water was ad libitum and environmental enrichment was provided. Animal welfare was monitored daily. Animals were randomly assigned to the experimental groups. Investigators were not blinded during the study. Experiments were performed in compliance with the Dutch legislation for the protection of animals used for scientific purposes (2014, implementing EU Directive 2010/63). Research was conducted under a project license from the Dutch competent authority (license number AVD1010020174312), and the study protocol was approved by the institutional Animal Welfare Body (Erasmus MC permit number 17-4312-22 and 17-4312-23).
Cells and viruses
Calu-3 cells were grown in Opti-MEM I Reduced Serum Medium (LIFE) supplemented with 10% FBS, penicillin (10,000 IU/ml) and streptomycin (10,000 IU/ml) in 5% CO2 at 37 °C. A passage 3 Bavpat-1 (D614G) (EVAg: 026V-03883, BetaCoV/Germany/BavPat1/2020 p.1) and a passage 3 Omicron BA.5 SARS-CoV-2 strain (EVAg: 010V-04723, hCovN19/Netherlands ZHNEMCN5892) grown and titrated on Calu-3 cells were used. Viral stocks were sequence confirmed.
Dose comparison
We evaluated two infectious doses for D614G and Omicron BA.5 in female (n = 12) and male (n = 12) hamsters. Virus dilutions for inoculation were prepared in Opti-MEM. Hamsters were inoculated intranasally with 103 or 104 tissue culture infectious dose-50 (TCID50) per animal in 20 μl (10 μl instilled dropwise in each nostril). Uninfected male (n = 6) and female (n = 4) control hamsters were age-matched to infected hamsters. Body weights were measured as a clinical parameter. Throat swabs (CLASSIQSwabs, Copan) were collected daily until 7 days post inoculation (DPI) (Supplementary Fig. 1A, B). Hamsters were euthanized by cervical dislocation under inhalation anesthesia (isoflurane) 7 DPI.
Determination of optimal co-housing duration
Donor hamsters (n = 6) were inoculated intranasally with 103 TCID50 per animal of Omicron BA.5 in 20 μl Opti-MEM (10 μl instilled dropwise in each nostril). 20 h post inoculation (HPI), each donor hamster was co-housed with 3 immunologically naïve recipient hamsters (n = 18). 6, 12 and 24 h post start of co-housing, one of the recipient hamsters was removed from each co-housing cage and re-housed to a new cage with recipient hamsters of the same co-housing time (Supplementary Fig. 2A). Body weights were measured as a clinical parameter. Throat swabs were collected daily until 14 DPI. Hamsters were euthanized by cervical dislocation under inhalation anesthesia (isoflurane) 14 DPI.
Comparative analysis of hamsters exposed via direct inoculation and direct-contact transmission
In a similar experiment, donor hamsters (n = 18) were inoculated intranasally with 103 TCID50 of Omicron BA.5 in 20 µl Opti-MEM (10 µl per nostril) and 20 h post-inoculation, each donor hamster was co-housed for 24 h with 3 immunologically naïve recipient animals (n = 10) (Supplementary Fig. 3A). On day 1, 2, 4 and 7 post-exposure (DPE; exposure for donor hamsters = moment of inoculation; exposure for recipient animals = end of co-housing) and at the end of the experiment (EOE; 14 DPE for donor and 12 DPE for recipient), donor (n = 4 per timepoint) and recipient (n = 2 per timepoint) hamsters were sacrificed and the head, lungs, trachea and nasal turbinates were harvested. Body weights were measured as a clinical parameter. Throat swabs were collected daily until the necropsy. Hamsters were euthanized by cervical dislocation under inhalation anesthesia (isoflurane) 14 DPI.
Sample processing
Throat swabs were stored at −80 °C in 250 µl virus transport medium (VTM; Minimum Essential Medium Eagle with Hank’s BSS (Lonza), 5 g L−1 lactalbumine enzymatic hydrolysate, 10% glycerol (Sigma-Aldrich), 200 U/ml of penicillin, 200 mg/ml of streptomycin, 100 U/ml of polymyxin B sulfate (Sigma-Aldrich), and 250 mg/ml of gentamicin (Life Technologies)). Blood was collected by cardiac puncture in serum-separating tubes (Greiner), processed, heat-inactivated and sera were stored at −80 °C. The lungs, trachea and nasal turbinates were harvested and a piece was transferred to 500 µl VTM. Tissues were homogenized with a tissuelyzer for 5 min at 30 Hz. The homogenate was centrifuged for 5 min at 4000 rpm. 60 µl of tissue homogenates or throat swabs in VTM were transferred to 90 µl lysis buffer (MagNA Pure External lysis buffer, Roche). The remaining homogenate was used without a freeze–thaw cycle for titrations. Remaining parts of the lung and half of the sagittal cut head were stored for 3 weeks in 4% neutral buffered formaldehyde before further processing.
RNA isolation and RT-qPCR on throat swabs and tissue homogenates
RNA isolation and RT-qPCR were performed as described previously25. Briefly, phocine distemper virus (PDV) was added as an internal control for the RNA extraction26 and the mixture was incubated with magnetic beads (AMPure XP, Beckman Coulter). After placement on a magnetic block (DynaMag™-96 Side Skirted Magnet (Thermo Fisher Scientific)), plates were washed three times with 70% ethanol. Beads were air-dried at room temperature and RNA was eluted in PCR grade water. RT-qPCR with the isolated RNA was performed using primers and probes targeting the E gene of SARS-CoV-2 and primers and probe for PDV detection26,27. Ct values of all samples were compared to a standard curve derived from a titrated Omicron BA.5 virus stock and TCID50 equivalents were calculated.
Virus isolation from throat swabs
SARS-CoV-2 was isolated from tissue- and throat-swab samples on Calu-3 using an infectious virus assay determining TCID50. Pre-diluted (1:2) throat swab samples or tissue homogenates were titrated on Calu-3 cells in a 3-fold dilution series in quadruplicate. Calu-3 cells were screened for cytopathic effect (CPE) after 6 days of incubation in 5% CO2 at 37 °C, and the infectious titer in TCID50/ml was calculated according to Reed and Muench28.
Detection of SARS-CoV-2-specific antibodies in hamster sera
Seroconversion was assessed in serum samples obtained during necropsy. Briefly, duplicates of heat-inactivated sera were two-fold diluted in Opti-MEM I Reduced Serum Medium supplemented with penicillin (10,000 IU/ml) and streptomycin (10,000 IU/ml) starting at a dilution of 1:10 in 60 µl. 400 SARS-CoV-2 foci forming units were added to each well in 60 µl and incubated in 5% CO2 at 37°C for 1 h. Virus-antibody mixtures were transferred to Calu-3 cells and incubated for 8 h. Cells were fixed and foci were stained with primary polyclonal rabbit-anti-SARS-CoV nucleocapsid antibody (1:1000; 40143-T62 SinoBiological) and secondary goat anti-rabbit IgG-Alexa FluorPlus488 (1:1000; A32731 Invitrogen). Plates were imaged using the Amersham Typhoon and the number of positive cells was quantified using Image Quant. The dilution that yields 50% reduction of foci (FRNT50) compared with the infection control was estimated by determining the proportionate distance between two dilutions.
Histopathology and immunohistochemistry
Lungs, trachea and sagittal cut hamster heads were stored in 4% formaldehyde (Avantor) during necropsy. Tissues were embedded in paraffin; 4 µm sections were prepared. Slides were routinely stained for hematoxylin and eosin for histopathological analysis and consecutive slides were stained for SARS-CoV-2 NP as described previously29. Scoring methodology was previously described30. Slides of hamster heads were scanned with the NanoZoomer 2.0-HT slide scanner.
Statistical analysis
Statistical analysis was performed on the raw data of the respective complete data set using two-way ANOVA or Kruskal–Wallis ANOVA in GraphPad Prism 9.5.0.
Results
Hamsters inoculated with BA.5 lose less body weight compared to D614G inoculated hamsters
Given the importance of viral shedding in the context of transmission studies, we compared two infectious doses to achieve reproducible infection and viral shedding for D614G and Omicron BA.5. Groups of female and male hamsters were intranasally inoculated with 20 µl containing 103 TCID50 or 104 TCID50 (Supplementary Fig. 1A, B). For an initial conformation of productive infection of hamsters, seroconversion was assessed at 7 DPI; all hamsters seroconverted (Supplementary Fig. 1C, D). Male hamsters inoculated with 104 TCID50 of Omicron BA.5 had significant higher relative bodyweight compared to male hamsters inoculated with 103 TCID50 of Omicron BA.5 1 DPI. Otherwise, no significant difference in body weight loss was observed between both doses for either variant (Fig. 1A, B). Compared to animals inoculated with Omicron BA.5, hamsters inoculated with D614G lost more body weight.
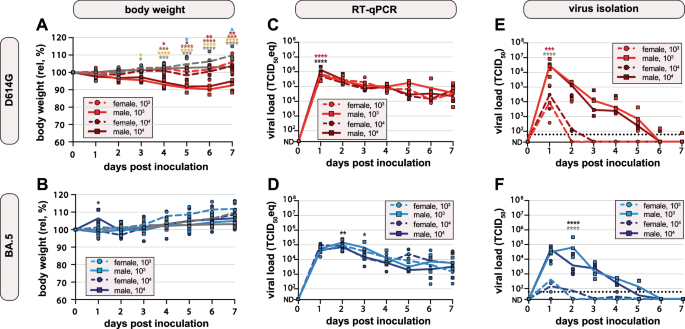
A, B Body weights were recorded daily. Relative body weights over time after A D614G or B Omicron BA.5 inoculation, day 0 was set at 100%. Symbols represent individual hamsters (n = 3 per group); line represents group means. Age- and sex-matched uninfected hamsters are shown in gray. C, D Viral load in throat swabs after C D614G or D Omicron BA.5 inoculation was measured by RT-qPCR and plotted as TCID50 eq/ml over time. Undetermined values (ND) were set as 20 TCID50 eq/ml. Symbols represent individual hamsters (n = 3 per group); line represents group geometric means. E, F Infectious virus in throat swabs after E D614G or F Omicron BA.5 inoculation was measured by virus isolation and plotted as TCID50/ml over time. The dotted lines represent the lower limit of detection (60). Undetermined values (ND) were set as 20 TCID50. Symbols represent individual hamsters (n = 3 per group); line represents group geometric means. Groups were compared using two-way ANOVA, corrected with Tukey. Asterisks indicate significant differences at specific timepoints and are shown in colors (female 103 vs. male 103: gray; female 104 vs. male 104: red; male 103 vs. male 104: black; male 103 vs. male uninfected: orange; male 104 vs. male uninfected: brown; female 104 vs. female uninfected: blue). *p < 0.05; **p < 0.01; ***p < 0.001.
Male hamsters lose more body weight than female hamsters
After inoculation with D614G, male hamsters lost approximately 10% of their body weight between 0 and 6 DPI. Male D614G infected hamsters had significantly lower body weight starting 3 DPI (103 TCID50 group) and 4 DPI (104 TCID50 group) compared to uninfected age-matched controls. Overall, the body weight of female hamsters remained stable. Female hamsters inoculated with 104 TCID50 of D614G had significantly lower body weight 5 DPI and 7 DPI compared to uninfected control animals. Relative body weights of female hamsters were significantly higher than those of male hamsters between 1 DPI and 7 DPI (103 TCID50 group) and between 4 DPI and 7 DPI (104 TCID50 group), indicating a sex-dependent effect of SARS-CoV-2 infections (Fig. 1A). Almost no body weight loss was observed in either female or male hamsters inoculated with Omicron BA.5, but female hamsters gained more weight compared to males (Fig. 1B). There was no significant difference in body weight of Omicron BA.5 inoculated hamsters compared to uninfected age- and sex-matched hamsters.
Hamsters inoculated with BA.5 have lower viral genome loads compared to D614G inoculated hamsters
Throat swabs were obtained daily and viral replication kinetics were assessed by RT-qPCR and virus isolation. SARS-CoV-2 genomes were detected in all inoculated hamsters by RT-qPCR (Fig. 1C, D). Viral genomes in the throats peaked on day 1 after SARS-CoV-2 D614G inoculation (comparable for both doses) and declined until the end of the experiment on 7 DPI (Fig. 1C). Viral genome levels were significantly higher in D614G-inoculated male than in female hamsters 1 DPI (104 TCID50 group). Additionally, viral genome levels were significantly higher in male hamsters after inoculation with 104 TCID50 than with 103 TCID50 at 1 DPI. Viral genome levels of BA.5 were overall lower than those of D614G. After Omicron BA.5 inoculation, viral genomes peaked at 1 DPI (104 TCID50 group) or 2 DPI (103 TCID50 group) and declined towards the end of the study period (Fig. 1D). Male hamsters inoculated with 103 TCID50 of BA.5 had significantly higher viral genome levels 2 and 3 DPI than male hamsters inoculated with a 104 TCID50 of BA.5. No significant difference in viral genome levels was detected between male and female hamsters after inoculation with either dose during the following days.
Male hamsters shed more infectious D614G and BA.5 than female hamsters
Virus isolations were performed to determine the infectious viral load in the throats of hamsters. All hamsters inoculated with a 104 TCID50, either D614G or BA.5, shed infectious virus (Fig. 1E, F). For the 103 TCID50 group, all hamsters shed infectious virus except for 1 out of 3 females inoculated with BA.5. Similar to the RT-qPCR data, infectious virus shedding peaked on day 1 after D614G inoculation, and on day 1 or 2 after BA.5 inoculation. Shedding of infectious D614G was up to 100-fold higher than that of BA.5. Notable discrepancies between the measurement of virus genomes and infections virus levels were observed for two aspects: (1) viral clearance, and (2) difference between male and female hamsters. In contrast to the genome levels, infectious virus loads declined fast and were no longer detected at 7 DPI. Notably, based on virus isolation data, the virus replication kinetics differed strongly between male and female hamsters in all groups. Up to 1000-fold more infectious virus was shed by male hamsters, independent of dose and challenge virus. Taken together, hamsters can be reproducibly infected with 103 TCID50 and low volume inoculum of D614G or Omicron BA.5. These experimental conditions were used for subsequent experiments. Compared to D614G, Omicron BA.5 led to lower viral loads and lower and delayed shedding of infectious virus. Shedding of infectious virus was always significantly higher in male than in female hamsters, independent of variant.
24 h co-housing-time starting 20 HPI ensures consistent BA.5 direct-contact transmission
Based on the virus isolation data, male hamsters were used to establish the Omicron BA.5 transmission model. As this variant was circulating at the time of experiment, we performed the subsequent experiments exclusively with this variant. To determine the optimal co-housing duration leading to consistent direct-contact transmission, each inoculated donor hamster was co-housed with 3 naïve hamsters starting 20 HPI of donor hamsters. At 6, 12 or 24 h post co-housing start, recipient animals were moved from infection cages to clean isolators (Supplementary Fig. 2A).
Similar to the dose-comparison experiment, no body weight loss was observed in either donor or recipient hamsters (Supplementary Fig. 2B). All donor hamsters proved productively infected based on increasing viral loads detected by RT-qPCR and virus isolation from throat swabs. Viral loads peaked on day 2 and declined until the donor hamsters were sacrificed on day 12 and 13 (Fig. 2A, D). Donor hamsters did not transmit Omicron BA.5 to recipient hamsters when co-housed for 6 h, and only 1 out of 6 recipient hamsters was infected by transmission after 12 h of co-housing. The viral replication kinetics in this hamster were similar to those observed in the donor hamsters, but with a delayed peak on day 4 post co-housing (6 DPI). Despite similar viral kinetics to donor hamsters, the infected recipient hamster did not transmit to his cage mates in the subsequent two weeks. A 24 h co-housing period led to consistent transmission to recipient hamsters. In comparison to the donor hamsters, the peak of viral genome in the throat was delayed, on day 3 post co-housing (5 DPI). Similar to the donor hamsters, viral loads decreased until the end of the experiment (Fig. 2B, E). Viral loads calculated as area under the curve (AUC) observed in donor hamsters were significantly higher than in recipient hamsters of the 6 and 12 h co-housing groups, and viral loads observed in the 24 h co-housing groups were significantly higher than in the 6 h co-housing groups (Fig. 2C, F). All inoculated donor and recipient hamsters in which virus replication was detected seroconverted (Supplementary Fig. 2C).
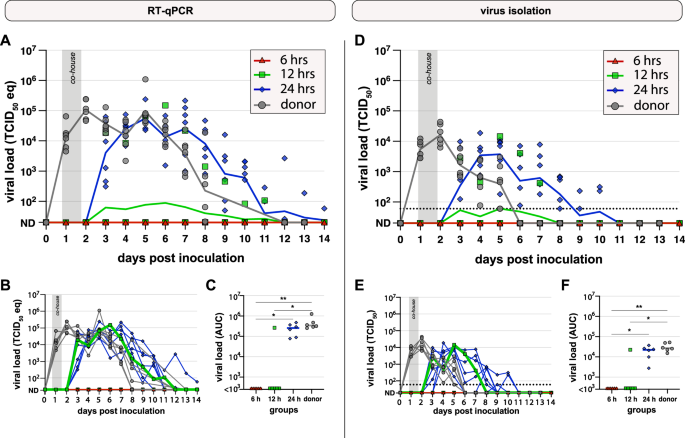
A–C Viral genome levels in throat swabs were measured by RT-qPCR. A Viral genomes plotted as TCID50 eq/ml over time after intranasal inoculation with Omicron BA.5 (donor, gray; n = 6) or co-housing with donor hamster for 6 h (red, triangle; n = 6), 12 h (green, square; n = 6) or 24 h (blue, diamond; n = 6) starting 20 h post inoculation of donors (HPI). Negative values (ND) were set to 20 TCID50 eq/ml. Symbols represent individual hamsters; lines represent group geometric means. B Identical to panel A, lines represent individual animals. C Area under the curve (AUC) based on the individual curves in panel B. Lines represent group median. Groups were compared by Kruskal–Wallis ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001. D–F Infectious viral loads in throat swabs were measured by virus isolation. D Infectious virus in throat swabs plotted as TCID50/ml. The dotted lines represent the lower limit of detection (60 TCID50/ml). Undetected values (ND) were set to 20 TCID50/ml. Symbols represent individual hamsters; line represents group geometric means. E Identical to panel D, lines represent individual animals. The dotted lines represent the lower limit of detection (60 TCID50/ml). F AUC based on the curves in panel E. Lines represent the group median. Groups were compared by Kruskal–Wallis ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001.
Viral loads in the respiratory tract are lower after direct-contact transmission when compared to direct inoculation
To characterize potential differences in pathogenesis after direct inoculation (donor hamsters) and infection by direct-contact transmission (recipient hamsters), we performed a virological and histopathological comparison between the respiratory tissues. To that end, Omicron BA.5-inoculated donor hamsters were co-housed with three immunologically naïve recipient hamsters for 24 h starting 20 HPI (Supplementary Fig. 3A). No body weight loss was observed in donor and recipient hamsters (Supplementary Fig. 3B). Viral loads in throat swabs as measured by RT-qPCR and virus isolation peaked on day 2 (donor) or day 3 (recipient) post exposure (DPE; donor exposure = inoculation, recipient exposure = co-housing end) and declined towards the end of the study period (Supplementary Fig. 3C, D). Donors (n = 4 per timepoint) and recipient (n = 2 per timepoint) were sacrificed on 1 DPE, 2 DPE, 4 DPE, 7 DPE and at the end of experiment (EOE).
The nasal turbinates from donor and recipient hamsters were RT-qPCR positive at all necropsy timepoints. Viral genome levels of donor hamsters peaked 2 DPE (geometric mean (gm): 1.8 × 107 TCID50eq/g tissue) and declined at later timepoints (EOE gm: 3.5 × 103 TCID50eq/g tissue). In comparison, viral genome loads in the nasal turbinates of recipient animals were low 2 DPE (gm: 1.5 × 103 TCID50eq/g tissue), peaked 7 DPE (gm: 3.1 × 105 TCID50eq/g tissue) and decreased until EOE (gm: 1.88 × 102 TCID50eq/g tissue) (Fig. 3A). Viral genome loads in the trachea of donor and recipient hamsters peaked 4 DPE with a gm of 1.5 × 105 TCID50eq/g tissue for donors and 4.5 × 103 TCID50eq/g tissue for recipients. At EOE, the viral genome loads in the trachea from donor and recipient hamsters decreased to low and undetectable RNA levels, respectively (Fig. 3B). Comparatively, low levels of RNA were detected in the lungs from donor and recipient hamster, which peaked 4 DPE (gm: 1.2 × 103 TCID50eq/g tissue) and 7 DPE (gm: 4.0 × 103 TCID50eq/g tissue), respectively, and declined to low and undetectable RNA levels at EOE (Fig. 3C). Infectious virus levels in the nasal turbinates of directly-inoculated donors were high at 1–4 DPE, and decreased to undetectable levels at EOE. In the nasal turbinates of the recipient hamsters, viral titers remained below the lower limit of detection, with the exception of low-level replication at 4 DPE (gm: 2.5 ×102 TCID50/g tissue) and 7 DPE (gm: 5.3 × 101 TCID50/g tissue) (Fig. 3D). In the trachea, infectious virus was detectable in all hamsters on 1-4 DPE, but with a higher titer for the donor hamsters compared to the recipients on all 4 days (peak gm: 2.1 × 104 TCID50/g tissue and 7.8 × 101 TCID50/g tissue, respectively) (Fig. 3E). Infectious virus was only detected in the lungs of donor hamsters 4 DPE at gm 9.6 × 101 TCID50/g tissue and no infectious virus was found in the lungs of recipient hamsters (Fig. 3F).
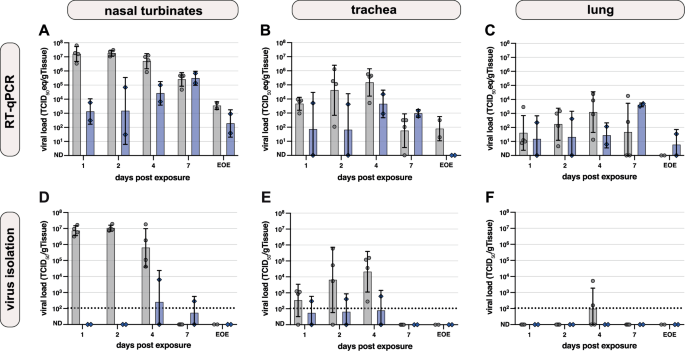
Donor hamsters are shown in gray (n = 4 per timepoint), recipient hamsters in blue (n = 2 per timepoint). Viral loads 1, 2, 3, 4, and 7 days after exposure (donor exposure = time of inoculation; recipient exposure = end of co-housing) and at end of experiment (EOE) are shown. A–C Viral genome loads in A nasal turbinate, B trachea and C lung after Omicron BA.5 inoculation measured by RT-qPCR and plotted as TCID50 eq/g tissue. Symbols represent individual hamsters, bars represent geometric mean and error bars show geometric SD. Undetected values (ND) were set to 1 TCID50 eq/g tissue (D–F). Infectious virus titers in D nasal turbinate, E trachea and F lung were determined by virus isolation and plotted as TCID50/g tissue. The dotted lines represent the lower limit of detection (106 TCID50/g tissue) based on the average tissue weight. Undetected values (ND) were set to 10 TCID50/g tissue. Symbols represent individual hamsters; bars represent geometric mean and error bars show geometric SD.
Taken together, we detected virus (RT-qPCR, virus isolation) in the upper respiratory tract of both donor and recipient hamsters, and to a lower extent in the lower respiratory tract. Viral genome loads were overall higher and peaked earlier in the donor hamsters compared to the recipients. Similarly, infectious virus levels were higher in the directly inoculated donors and were overall only detected in the first days after exposure.
Mild inflammation in the respiratory tract of Omicron BA.5 infected donor and recipient hamsters
Histopathological evaluation (Supplementary Table 1) of the nasal mucosae showed slight to mild inflammation in all donor and recipient hamsters from 2 DPE onwards. This rhinitis was evidenced by intra-epithelial infiltrates of neutrophils and degenerated to sloughed epithelial cells (Supplementary Fig. 4A), that co-localized with cytoplasmic positivity for SARS-CoV-2 antigen stained by immunohistochemistry (IHC). Despite the absence of inflammation at 1 DPE, SARS-CoV-2 antigen staining was also positive at that timepoint (Fig. 4). Antigen was predominantly detected in the respiratory epithelium of the nasal mucosa, whereas in the 2 and 4 DPE donor hamsters the nasal olfactory mucosa was also positive. Regardless of belonging to donor or recipient groups, nearly all hamsters showed faint ciliary to mild cytoplasmic viral positivity of tracheal mucosal epithelial cells, in conjunction with slight to mild concomitant inflammation consisting of infiltrating neutrophils only present in a few 4 and 7 DPE donor hamsters and in 7 DPE recipient hamsters (Supplementary Fig. 4B). Likewise, the majority of hamsters in both the donor and recipient groups showed mostly faint ciliary viral positivity of bronchial mucosal epithelial cells, devoid of inflammation. Minor chronic inflammatory consolidated alveolar foci in few lungs of 7 DPE donor and recipient animals were interpreted as background lesions since these, and all others hamster lungs, were devoid of viral positivity by IHC, as were all other organs examined. In line with viral loads detected by RT-qPCR and virus isolation, donor hamsters overall showed (slightly) more inflammation and higher antigen positivity compared to recipients (Supplementary Fig. 5). Taken together, histopathological evaluation indicated mild inflammation and viral antigen positivity in the upper respiratory tract and viral antigen negativity in the lower respiratory tract.
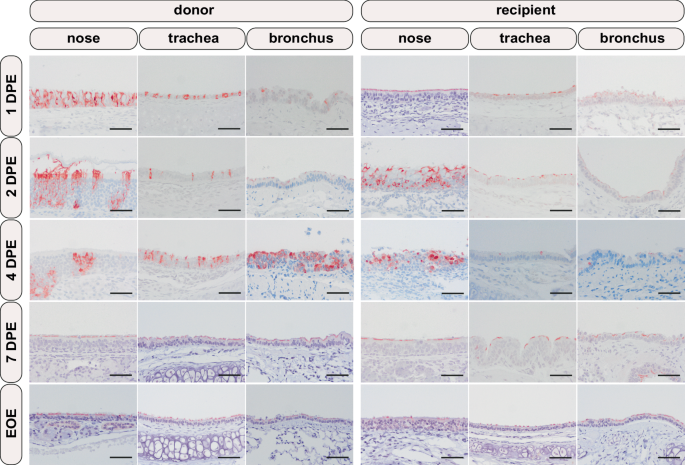
From left to right, nasal, tracheal, and bronchial mucosae are depicted. From top to bottom, the consecutive day’s post exposure (DPE; donor exposure = time of inoculation; recipient exposure = end of co-housing) and at the end of experiment (EOE; 14 DPE for donor, 12 DPE for recipient) are depicted. Positive viral antigen is visualized as reddish-brown staining of the epithelial cells’ cytoplasm and/or cilia by AEC-immunoperoxidase, on haematoxylin counterstain. Original magnifications 200×. Scale bar 100 µm.
Discussion
As SARS-CoV-2 continues to evolve antigenically to escape vaccine- or infection-induced immunity, suitable animal models are needed to study novel interventions against individual viral variants. In this study, we established an Omicron BA.5 direct-contact transmission model in hamsters. We compared (1) two doses of SARS-CoV-2 D614G and BA.5 for productive infection, determined (2) the optimal hamster sex, and (3) optimal co-housing timing and duration to achieve reproducible direct-contact transmission. Furthermore, we showed (4) differences in the histopathology and viral load in the respiratory tract of donor and recipient hamsters.
Different challenge doses of SARS-CoV-2 for intranasal inoculation of hamsters have been described before, ranging from 100 to 106 TCID50. Commonly, an intranasal infectious dose of approximately 105 TCID50 is used22,31,32,33,34,35,36,37,38. Omicron BA.1 and BA.2 have shown reduced pathogenicity and transmission potential compared to earlier variants24. We assumed that a higher dose compared to D614G might be necessary for Omicron BA.5 to ensure robust infection and transmission. To determine an infectious dose of SARS-CoV-2 D614G and Omicron BA.5 needed to ensure productive infection, we compared infectious doses of 103 TCID50 and 104 TCID50. Intra-nasal inoculation of 103 TCID50 per animal was sufficient to infect 6 out of 6 hamsters. Despite all hamsters being SARS-CoV-2 positive with comparable viral loads measured by RT-qPCR, male hamsters shed higher loads of infectious virus than female hamsters. At the same time, male hamsters showed higher weight loss than females, demonstrating an association between infectious virus loads and clinical signs. Sex-related differences in hamsters inoculated with SARS-CoV-2, including histological lesions, antibody responses and infectious virus loads have been described before and reflect disease severity in humans suffering from COVID-19. SARS-CoV-2 infected male hamsters lost more body weight after infection and despite detection of comparable viral genome loads, infectious virus titers are higher in male hamsters39,40,41,42,43. It has been suggested that the interaction of SARS-CoV-2 spike protein with estrogen receptors could (partly) be responsible for these findings42. It would be beneficial to investigate sex-dependent differences in future studies. Importantly, the higher shedding of infectious virus makes male hamsters more suitable for use in transmission studies for testing intervention strategies.
Different volumes have been used for intranasal inoculation of hamsters, ranging from 20 to 200 µl31,34,35,44,45,46,47,48,49. It has been shown that disease severity is influenced by the volume of the inoculum, even when given without an infectious agent50,51. Intranasal inoculation with a high volume of inoculum can lead to direct deposition of SARS-CoV-2 into the lower respiratory tract. The amount of inflammation and clinical signs increases with the volume50,52,53,54,55. To ensure deposition and the start of replication in the upper respiratory tract, we used a small volume inoculum of 10 µl per nostril (20 µl per hamster). This closely resembles the preferential initial replication of Omicron in the upper respiratory tract of humans56. Indeed, we detected almost no viral antigen or inflammation in the lungs of directly inoculated hamsters, and there was almost no infectious virus detectable. Viral loads detected by RT-qPCR increased in the first days after inoculation. This indicates that with a small volume the infection starts in the upper respiratory tract and spreads from there.
In throat swabs taken from male hamsters inoculated with D614G or Omicron BA.5, we detected infectious virus until 6 to 7 DPI with a peak in the first days post inoculation. Viral shedding of SARS-CoV-2 in hamsters has been described in literature between 1 and 14 DPI57. We show that the viral loads in throat swabs of Omicron BA.5 inoculated hamsters were lower compared to D614G, and that Omicron BA.5 infection led to less body weight loss3. Similar, it has been reported before that viral loads are lower in Omicron BA.1 inoculated hamsters compared the earlier variant Delta. Due to the demonstrated differences in viral kinetics, shedding and clinical signs, there is a need for well-characterized animal models for the individual variants of SARS-CoV-2. Previous studies have shown that directly inoculated donor hamsters efficiently transmit variants of SARS-CoV-2, such as the Beta or Delta variant, to naïve hamsters by direct-contact, fomites or via the air3,22,58. During a direct-contact transmission study, not only air-borne transmission but also fomite transmission plays a role. However, it has been described that fomite transmission efficiency is lower compared to air-borne58. An intervention that protects from direct-contact transmission should also protect from air-borne transmission. Therefore, we developed a direct-contact transmission model.
At the time of performing this study, Omicron BA.5 was the circulating variant and therefore, we established the direct-contact transmission model for this virus. The higher infectious virus titers in male hamsters promised better transmission to recipient hamsters and can serve as an evaluation criterion for future intervention studies. Direct-contact transmission of SARS-CoV-2 has been demonstrated before, but to our knowledge no Omicron BA.5 transmission model has been characterized. For the Beta variant, intranasal challenge of donors with 8 × 104 TCID50 in 80 µl and co-housing start at 24 HPI resulted in 100% direct-contact transmission to naïve recipients22. When donor hamsters were inoculated with 105 TCID50 Omicron BA.1 in 100 µl, and co-housed with naïve recipients for 4 h starting 24 HPI, 3 out of 3 recipient hamsters were positive3. Here, we show that male hamsters inoculated with 103 or 104 TCID50 of Omicron BA.5 displayed a peak in shedding between 1 and 2 DPI. Since transmission to immunologically naïve recipient animals is optimal when shedding is highest59, we started co-housing of donor and recipients 20 HPI and stopped 6, 12, or 24 h later. Only in the 24 h-co-housing group, reproducible direct-contact transmission was established. In the 12 h group only one hamster, and in the 6 h co-housing group no recipient animals turned positive. Compared to earlier variants of SARS-CoV-2, the co-housing time for Omicron BA.5 is longer. This seems to differ from the human situation, where Omicron BA.5 transmitted more efficiently and outcompeted earlier variants60. However, it is important that transmission dynamics in humans are based on an immune population and not naïve individuals. Further, these results indicate a co-housing-time dependence of effective transmission, but we cannot exclude that not the duration of co-housing, but the time of co-housing initiation is crucial. A shorter co-housing period could be enough if precisely at time of highest shedding. Further, hamsters are nocturnal in captivity and it might be necessary that cohousing is (partly) during the night, when hamsters show most activity and are expected to interact and transmit the most61. Due to practical reasons, we started co-housing in the morning. Further optimization could be done, but 24 h co-housing starting at 20 HPI are robust choices to ensure reproducible direct-contact transmission. We performed transmission experiments with hamsters that were 5–6 weeks old. 4–8-week-old hamsters are commonly used in SARS-CoV-2 studies22,62,63,64. We cannot exclude that outcomes differ for aged hamsters.
Shedding of infectious Omicron BA.5 virus was detected in both donor and recipient hamsters, but infectious titers peaked later and were lower in recipient hamsters. A similar decrease in magnitude of shedding and delay of peak was also described in an airborne transmission hamster model for SARS-CoV-2 Alpha and Delta32. It was shown that the percentage of hamsters with virus positive throat swabs decreases with increasing rounds of transmission. While throat swabs of 8 out of 8 primary donors and recipients were infectious virus positive, this decreased to 4 out of 8, and 1 out of 8 in consecutive transmission rounds32. Similarly, in our study, one recipient hamster of the 12 h co-housing group got infected, but did not spread further to his cage mates. Infectious titers in throat swabs were comparable to donor hamsters but peaked lower.
To further characterize the differences in pathogenesis between donor and recipients, we compared viral load in organs of the respiratory tract and performed histopathological analysis of the respiratory tract. Overall, viral loads in throat swabs detected by RT-qPCR and titration peaked delayed in recipient compared to donor hamsters. Viral loads were lower and peaked later in the nasal turbinate, trachea and lung of recipient animals compared to donor hamsters. Particularly, viral loads detected in the nasal turbinate 1 DPE differed strongly between donors and recipients. We speculate that the challenge dose transmitted from donors to recipients by direct-contact is lower than the dose used for direct-inoculation of donors. Therefore, viral loads in the nasal turbinates of recipient hamsters were lower directly post exposure, spread to the rest of the respiratory tract was slower and infectious titers peaked later. Histopathological results show a productive virus infection and subsequent inflammation confined to the upper respiratory tract in both the inoculated donor hamsters as well as in the exposed recipient hamsters. The induced rhinitis was mild to moderate, the tracheitis was mild, and inflammation—despite faint viral positivity by IHC—was absent in the bronchi. These findings corroborate the other virological results of an efficacious viral infection, excretion and transmission in this model.
Taking the differences described above into account, combined with the fact that viral exposure of hamsters by contact with infected donors mimics exposure in humans, we conclude that testing interventions in hamster transmission models could lead to results that are more readily translated to humans, compared to direct-inoculation models. Emergence of novel SARS-CoV-2 variants with different replication kinetics emphasizes that newly characterized models are required for each variant. Here, we established a robust Omicron BA.5 direct-contact transmission model that can be utilized to study prophylactic and therapeutic interventions against SARS-CoV-2.


Responses