Condensate remodeling reorganizes innate SS18 in synovial sarcomagenesis

Introduction
Synovial sarcoma, a rare and relatively malignant soft tissue sarcoma, constitutes ~7% of all soft tissue sarcomas, with an incidence of around 1 in 500,000 [1]. More than 95% of synovial sarcoma patients exhibit a characteristic translocation of chromosome 18 with chromosome X, leading to the fusion of SS18 with SSX1, SSX2 or SSX4 and the formation of the oncogenic fusion protein SS18-SSX [1, 2]. The SS18-SSX fusion protein retains the N-terminal domain and QPGY domain of SS18, with only 8 amino acids at the SS18 C-terminal are replaced by 78 amino acids at the C-terminal of SSX [3, 4].
It has been shown that the oncogenic fusion protein SS18-SSX hijacks the BAF complex from the enhancer region to the broad polycomb domain, thereby aberrantly activating a large number of genes such as SOX2, PAX7, CAV1, and SOX8 [3, 5]. It also has been discovered that the PRC1.1 complex containing KDM2B could promote the recruitment of the SWI/SNF complex, where SS18-SSX1 is located, to unmethylated CpG islands, thereby increasing gene accessibility and leading to abnormal activation of developmental genes [6]. Further, the oncogenic fusion protein SS18-SSX were found to exhibit a strong preference for H2AK119ub-labeled nucleosomes [7, 8]. The tumorigenic mechanism of the fusion protein SS18-SSX mainly focuses on the regulation of chromatin remodeling complexes by SS18-SSX, altering protein interactions to enhance its oncogenic activity. From this perspective, researchers have found that some small molecules can indirectly affect the tumorigenicity of SS18-SSX [9, 10]. However, these small molecules lack specificity for the SS18-SSX. Given this challenge, a fresh perspective is needed to delve into the study of the SS18-SSX.
Currently, phase separation-related research has penetrated into a large number of fields of biological research. Such as gene expression regulation [11, 12], DNA damage repair and chromatin structure remodeling [13, 14], cell junction [15], autophagy [16], signaling pathway regulation [17], cell fate transition [18, 19], etc. In recent years, researchers have found that phase separation also plays an important role in the development of tumorigenesis. For example, the leukemia-associated oncogenic fusion protein NUP98-HOXA9 contains a phenylalanine (FG) repeat sequence that enables it to undergo liquid-liquid phase separation (LLPS), in which the NUP98-HOXA9 mutant that loses LLPS lacks tumorigenic activity when expressed in mouse hematopoietic stem cells and progenitor cells [20, 21]. These suggest that the abnormal acquisition or loss of LLPS can fundamentally change cellular homeostasis and drive tumorigenesis, providing new insights for understanding the mechanism of carcinogenesis.
Previously, our laboratory discovered that the QPGY domain of SS18 protein is an intrinsic disordered region (IDR), conferring upon the SS18 protein the capability of phase separation. Notably, the phase separation is disrupted when tyrosine is mutated to alanine, and at the same time, the regulatory ability of SS18 in the process of pluripotency to somatic cell conversion is also changed [18]. Intriguingly, it has been observed that the oncogenic fusion protein SS18-SSX also exhibits phase separation ability dependent on the tyrosine-rich QPGY domain [22, 23]. However, it remains unclear whether there are differences in biological functions and physical properties between the condensates formed by SS18 and SS18-SSX. Exploring the properties of SS18-SSX protein condensates may offer a novel direction for the treatment of synovial sarcoma.
Results
SS18/SS18-SSX reveal distinct properties in nuclei
Patients with synovial sarcoma will have chromosome 18 and X chromosome translocations, leading to replacing the C-terminal 8 amino acids of wild-type SS18 by C-terminal 78 amino acids of SSX (SSX1, SSX2 or SSX4) to form oncoprotein SS18-SSX (Fig. 1A). To understand the differences between SS18 and SS18-SSX, we first analyzed their condensates by overexpression in BJ cells. We observed a significant increase in the number of SS18-SSX condensates, which was ~150% more efficient than SS18 (Fig. 1B, C). Further, we examined the distribution of SS18 protein and SS18-SSX1 protein in synovial sarcoma HS-SY-II cells expressing SS18-SSX1 protein. Since the SS18-SSX protein retains the epitope of SS18 antibody, to examine the distribution of SS18 protein in HS-SY-II cells, we knock out the SS18-SSX1 protein (Fig. S1C). It shows that the number of endogenous condensates of SS18 protein and SS18-SSX1 protein in HS-SY-II cells are too excessive to count (Fig. 1D), thus we attempt to analyze the diameter of the condensates. We found endogenous SS18-SSX1 condensate diameter significantly larger than SS18 in the synovial sarcoma cell line HS-SY-II cells (Fig. 1E). We then tested their sensitivity to 1,6-hexanediol (1,6-Hex), an aliphatic alcohol capable of disrupting weak hydrophobic interactions [24] and show that 3% 1,6-Hex could disrupt the formation of SS18 condensates within 30 s (Fig. 1F), but has little effect on SS18-SSX condensates (Fig. 1G). It took 20% 1,6-Hex to dissolve SS18-SSX condensates in 30 s (Fig. 1G). Furthermore, we performed fluorescence recovery after photobleaching (FRAP) experiments to analyze the diffusion kinetics of these two condensates and show that the molecular exchange velocity of SS18-SSX onco-condensate is significantly lower than that of SS18 (Fig. 1H, I). Consistently, we show that SS18-SSX condensates are more temperature resistant than SS18 as it takes up to 42 °C to dissolve SS18-SSX condensates in 30 min, compared to 40 °C in 2 min for SS18 (Fig. S1A, B). These results suggest that SSX C-terminal fusion transforms an innate condensate into an oncogenic one by remodeling their physical properties, and SS18-SSX onco-fusion protein has a stronger tendency to form phase separation than innate SS18 in nuclei.
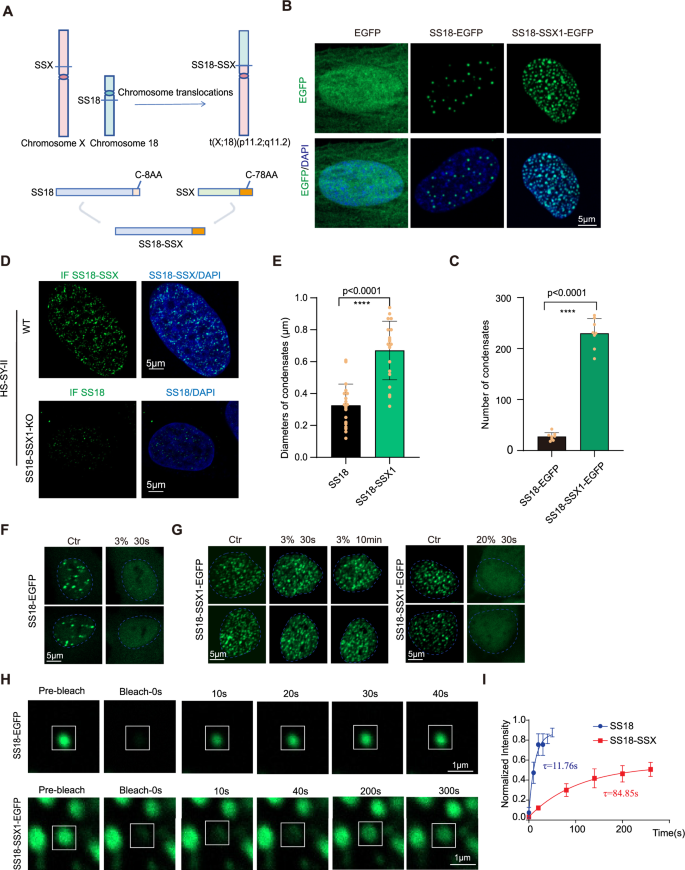
A Schematic illustration for SS18-SSX onco-fusion in synovial sarcoma. The C-terminal 8 amino acids of SS18 were replaced by 78 amino acids of SSX C terminus. B Representative images of EGFP, SS18-EGFP and SS18-SSX1-EGFP fusion protein overexpressed via lentiviral infection in BJ fibroblasts. Scale bars, 5 μm. C Histogram for the number of condensates quantified from (B). Data are mean±s.d., two-sided, unpaired t test; n = 8 nuclei. D The upper row shows representative images of immunofluorescence of the endogenous SS18-SSX1 protein in the HS-SY-II cell line, and the bottom row shows representative images of immunofluorescence of the endogenous SS18 protein after SS18-SSX1 protein knockout in HS-SY-II. Scale bars, 5 μm. E Histogram for the diameters of condensates quantified from (D). Data are mean±s.d., two-sided, unpaired t test; n = 20 condensates. F Representative images of BJ fibroblasts expressing SS18-EGFP when treated with 3% 1.6-hexanediol (1.6-hex). The blue dotted line represents the nuclear boundary. Scale bars, 5 μm. G Representative image of BJ fibroblasts expressing SS18-SSX1-EGFP when treated with 3% or 20% 1.6-hex. The blue dotted line represents the nuclear boundary. Scale bars, 5 μm. H Representative image of FRAP experiments with lentiviral SS18-EGFP or SS18-SSX1-EGFP expressed in BJ fibroblasts. The condensates undergone bleaching are indicated in the white box. The white line represents the condensate boundary. Scale bars, 5 μm. I Quantification of SS18-EGFP and SS18-SSX-EGFP in FRAP experiments in (H). The intensity dynamics of the bleached regions of SS18 or SS18-SSX1 condensates were fit to an exponential function, respectively. τ(SS18) = 11.76 s, τ(SS18-SSX) = 84.85 s; Data are mean±s.d., two-sided, unpaired t test; n = 6 condensates.
H2AK119ub recognition impels condensate remodeling
Our results described above are in sharp conflict with those reported in vitro, i.e., SS18-SSX has weaker LLPS ability than SS18 [22]. We hypothesize that the nuclear environment plays a role in transforming a seemingly weaker condensate into a much stronger one. To test this hypothesis, we performed small-molecule screening to identify compounds that could disturb the formation of SS18-SSX condensates (Fig. 2A). We used lentivirus to infect BJ cells to express the fusion protein SS18-SSX1-EGFP. Five days later, we conducted small molecule drug screening and screened a total of 6500 small molecules. We were surprised to find a small molecule PRT4165 that could diffuse SS18-SSX1 condensates (Fig. S2A). It is worth mentioning that this dispersion is not reflected in all cells, but in some cells. We identified PRT4165, a potent inhibitor of PRC1-mediated H2A ubiquitylation [25], that disrupt the SS18-SSX condensates and has little effect to that of SS18 (Fig. 2B, C). Consistently, inhibition of H2AK119ub by PRT4165 could also restrain the tumorigenicity of SS18-SSX1 as tested by EdU cell proliferation assay (Fig. S2B, C). This suggests that remodeling of the oncogenic fusion protein SS18-SSX condensates is associated with H2AK119ub in cells compared with wild-type SS18.
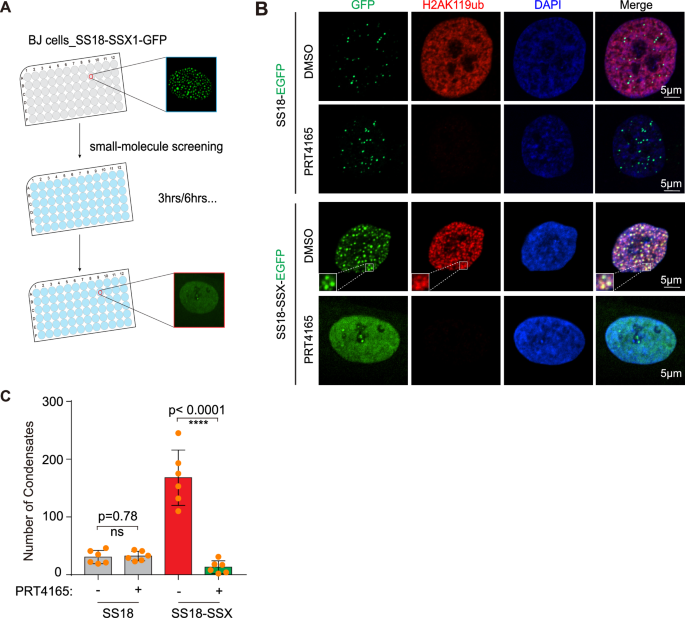
A Schematic of workflow for screening small molecules capable of disrupting SS18-SSX condensates. B Representative images of BJ fibroblasts expressing SS18-EGFP and SS18-SSX1-EGFP for immunofluorescence detection of H2AK119ub after 3 h treatment with DMSO and 37.5 μM PRT4165. AF568 (red light) uses the same exposure time of 100 ms, scale bar, 5 μm. C Histogram with the number of condensates in (B). Data are mean±s.d., two-sided, unpaired t test; n = 6 nuclei. ****P < 0.0001.
To further explore the connection between SS18-SSX condensate and H2AK119ub, we tested the chromatin binding of each protein and show that they overlap very little (Fig. S3A). Instead, SS18-SSX, not SS18, binds to sites marked by H2AK119ub (Fig. S3B, D), but the relocation did not significantly affect the original H2AK119ub (Fig. S3C). This indicates that the SS18-SSX1 protein does not change the H2AK119ub modification site, but prefers to bind to the H2AK119ub modification site.
The formation of SS18-SSX condensates relies on the acidic region of C-SSX
We further show that H2AK119ub mediates SS18-SSX condensation through an acidic region (AR) of C-SSX (Fig. 3A), which was reported to interact with H2AK119ub [7]. We created the AR mutant of SS18-SSX with the acidic region residues substituted by alanine and show that the acidic region mutant (AR Mut) of SS18-SSX is no longer able to form condensates (Fig. 3B, C) and fails to bind the histone H2AK119ub modification sites (Fig. 3D). Further, we see that SS18-SSX (AR Mut) can not activate the expression of the downstream genes (Fig. 3E) and also lost the ability to stimulate cell proliferation, then presumably tumorigenicity (Fig. 3F, G). These results indicate that the formation of SS18-SSX condensates depends on the binding of the acidic region of C-SSX to the widely distributed histone modification H2AK119ub.
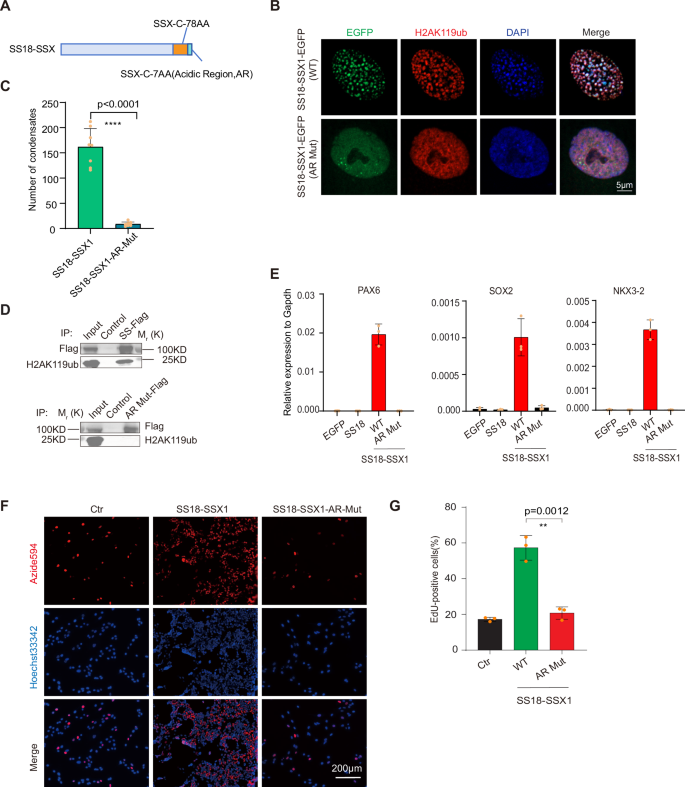
A Schematic illustration for SS18-SSX onco-fusion protein. The C-terminal acidic region (AR) DPEEDDE → AAAAAAA was highlighted. B Representative image of immunofluorescence for H2AK119ub in BJ fibroblasts with the lentiviral expression of wild-type or AR mutant SS18-SSX1-EGFP. Scale bars, 5 μm. C Histogram shows the number of condensates in BJ fibroblasts with lentiviral expression of wild-type or AR mutant SS18-SSX1-EGFP in (B). Data are mean±s.d., two-sided, unpaired t test; n = 8 nuclei. D Pull-down assays of the SS18-SSX1-EGFP-Flag (SS-Flag) and SS18-SSX1(AR Mut)-EGFP-Flag (AR Mut-Flag) protein. Binding to the H2AK119ub was shown by immunoblotting. E The expression of representative downstream genes of SS18-SSX in BJ fibroblasts with lentiviral expression of EGFP, SS18, wild-type and AR mutant SS18-SSX1. Data are mean±s.d., n = 3 independent experiments. F Representative image of EdU assay in synovial sarcoma cell line SW982 with the lentiviral expression of EGFP, wild-type and AR mutant SS18-SSX1. Scale bars, 200 μm. G Histogram shows the ratio of EdU-positive cells in (E). Data are mean±s.d., two-sided, unpaired t test, **P < 0.01. n = 3 independent experiments.
SS18 relocates in a phase separation manner
As translocation occurs in only one set of chromosomes, wild-type SS18 and oncogenic SS18-SSX should be expressed and coexist within one cell. We wish to test how these two different condensates interact with each other (Fig. 4A). Remarkably, we show that both wild-type SS18 and oncogenic SS18-SSX co-localize in the same condensates (Fig. 4B). Moreover, there are also a large number of H2AK119ub modifications on the condensates formed by SS18 (Fig. S4A). However, in BJ cells overexpressing SS18 alone, the SS18 protein site does not have H2AK119ub modifications (Fig. S4A). Remarkably, SS18 now occupies chromatin loci occupied by SS18-SSX, but no longer binds to those it normally does (Fig. S4B, C). As a result, this relocation or hijacking of SS18 leads to dramatic downregulation of SS18 targets such as FGF2 and tumor suppressor gene DAB2, CAV1 (Fig. 4C, D), These results suggest that SS18-SSX is dominant over SS18 through preferential condensations.
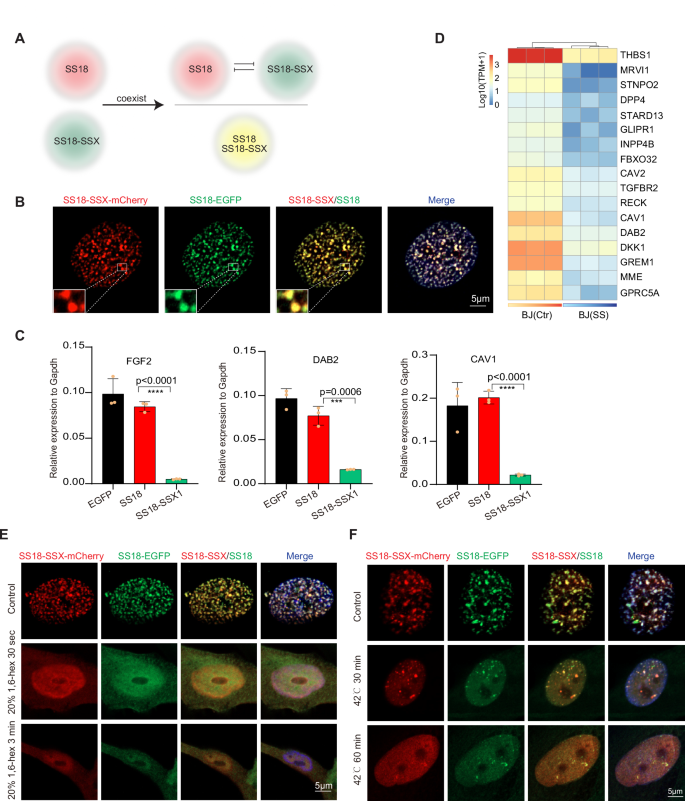
A Schematic illustration indicates the competition between endogenous SS18 condensates and pathological SS18-SSX condensates coexisting within synovial sarcoma cells. B Representative images of immunofluorescence in the BJ fibroblasts with lentiviral co-expression of SS18-SSX1-mCherry and SS18-EGFP. Scale bars, 5 μm. C The expression of representative downregulated genes in SS18-SSX expression. Data are mean±s.d., two-sided, unpaired t test, ***P < 0.001, ****P < 0.0001. n = 3 independent experiments. D Heatmap shows the expression of representative downregulated tumor suppressor genes upon SS18-SSX expression in BJ fibroblasts. E Representative images of immunofluorescence in the BJ fibroblasts with lentiviral co-expression of SS18-SSX1-mCherry and SS18-EGFP after 20% 1,6-hex treatment for 30 s or 3 min. Scale bars, 5 μm. F Representative images of immunofluorescence in the BJ fibroblasts with lentiviral co-expression of SS18-SSX1-mCherry and SS18-EGFP after incubated at 42 °C for 30 min or 1 h. Scale bars, 5 μm.
Since SS18-SSX1 condensates can be diffused with the treatment of 1,6-hex or high-temperature conditions, we next attempted to destroy SS18-SSX1 condensates and release SS18 condensates to rescue the expression of FGF2, CAV1, and DAB2, which are targeted by SS18 protein. We co-overexpressed the SS18-EGFP and SS18-SSX1-mCherry proteins within BJ cells, which were later treated with 1,6-hex and high temperature. It shows that 20% 1,6-hex can make the SS18-SSX1-mCherry condensates diffused, meanwhile, the SS18-EGFP condensates also become diffuse (Fig. 4E). After incubation at 42 °C for 30 min or 1 h, the number of the SS18-SSX1-mCherry condensates is significantly reduced, but the SS18-EGFP protein condensates are still present in the SS18-SSX1-mCherry protein condensates (Fig. 4F).
Furthermore, we found that the FGF2, CAV1 and DAB2 expression levels were increased after the treatment with 20% 1,6-hex for 3 min, as compared with the control group (Fig. S5A), whereas FGF2, CAV1, and DAB2 expression levels were not significantly increased in cells cultured at 42 °C for 30 min or 1 h (Fig. S5B). These data suggest that the phase separation mechanism mediated SS18 relocation serves a significant role in gene regulation.
Discussion
Previously, we revealed that SS18 can undergo phase separation [18]. In this study, we identified that C-SSX affects the physical properties of wild-type SS18 condensate. The oncogenic fusion protein SS18-SSX is significantly different from the proto-oncoprotein SS18 in four aspects: First, the number of SS18-SSX protein condensates is significantly increased; second, SS18-SSX condensate is more resistant to 1,6-Hex; third, the two types of condensates present distinct molecular dynamics; lastly, onco-fusion condensate is more temperature resistant.
To explore the mechanism underlying the changes in the physical properties of the intracellular fusion protein SS18-SSX1 condensates, we performed a drug screening and found that the inhibitor of H2AK119ub histone modification PRT4165 can diffuse the condensates in the nucleus of BJ cells expressing SS18-SSX1-EGFP, significantly. Importantly, downstream genes, such as SOX2, PAX7, and NKX3-2, are no longer activated by the condensate-altered SS18-SSX (AR Mut). Meanwhile, SS18-SSX (AR Mut) no longer promotes cell proliferation compared to wild-type SS18-SSX. This alteration of SS18-SSX1 condensates is dependent on H2AK119ub recognition by C-SSX. Further, we found that the oncogenic fusion protein SS18-SSX1 condensate recruited SS18 protein condensate, changed the localization of SS18 protein, and impeded the expression of SS18-targeted tumor suppressor genes (Fig. 5). These data indicate that the competition between different condensates could be a new dimension to dominate cell fate.
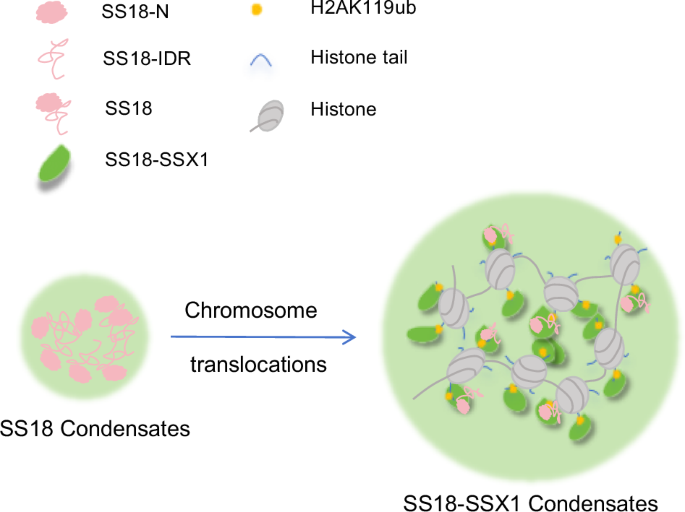
Schematic diagram of wild-type SS18 protein condensates formation (left panel). Schematic diagram of oncogenic fusion protein SS18-SSX condensates formation (right panel). The formation of SS18 protein condensates depended on the IDR sequence, and the SS18-SSX-enhanced condensates relied on H2AK119ub. Significantly, the SS18-SSX condensates recruit the proto-oncogene protein SS18.
Many soft tissue sarcomas, such as myxoid liposarcoma, Ewing sarcoma, extraosseous myxoid chondrosarcoma, etc., are also caused by chromosomal translocations [26, 27]. Studies have shown that these fusion proteins can also form condensates [28]. This suggests that the occurrence of these sarcomas may also be related to the aberrant condensate. Our works and condensate remodeling paradigm could provide new ideas for studying these related diseases.
Materials and methods
Cell culture
HEK293T cells (ATCC, CRL-1126) and SW982 cells (ATCC, HTB-93) are cultured in DMEM supplemented with 15% FBS, GlutaMAX and NEAA. BJ fibroblasts (MEISEN CELL, CTCC-400-0144) are cultured in DMEM/F12 supplemented with 15% FBS, GlutaMAX, and NEAA. The synovial sarcoma cell line HS-SY-II was purchased RIKEN BRC (RCB22316). HS-SY-II cell line was cultured using standard protocols in DMEM low glucose medium (Gibco) supplemented with 10% FBS. Each cell line was grown in a humidified incubator at 37 °C with 5% CO2. We used PRT4165 (TargetMol T3110) to block the H2AK119ub modification. All the cell lines have been confirmed as mycoplasma-free with Beyotime C0297M.
Stable gene expression and gene knockout sgRNA constructs
Constitutive expression of SS18, SS18-HA, SS18-SSX1 and SS18-SSX1 AR-mutations was obtained using an EF1alpha-driven expression vector (pKD-EF-1a-MCS-EGFP/mCherry-IRES2-Puro or pKD-EF-1a-MCS-EGFP/mCherry-IRES2-Blast) expressed in cells by lentiviral infection and selected with puromycin (2 μg/ml) or blasticidin (10 µg/ml) after 48 h. Constitutive expression of sgRNA targeting the region of C-SSX of the SS18-SSX fusion (5′-gCATGCCCAAGAAGCCAGCAG-3′) or a NT non-targeting control (5′-GGTAGCGAACGTGTCCGGCGT-3′) was obtained using lentiviral infection of the Lenti-Cas9 -Blast vector with blasticidin (2 μg/ml) selection.
Lentivirus generation
Lentivirus production was obtained from PEI (Polysciences, #23966) transfection of HEK293T cells with co-transfection of the packaging vectors pSPAX2 and pMD2.G along with the gene delivery vector. Viral supernatants were collected 48 h after transfection. For infection, the viral pellets were added to cells in a dropwise manner in the presence of polybrene. After 8 h, medium containing the lentivirus was replaced. Infected cells were selected by the addition of puromycin (2 μg/ml) or blasticidin (10 µg/ml) after 48 h.
qRT–PCR
Total RNAs were prepared with TRIzol. For quantitative PCR, cDNAs were synthesized with ReverTra Ace (Toyobo) and oligo-dT (Takara), and then analyzed by qPCR with ChamQ SYBR qPCR Master Mix (Vazyme). The qPCR primer sequences are listed in Supplementary Table S1.
Immunofluorescence
Cells growing on coverslips were washed three times with PBS, then fixed with 4% PFA for 30 min, and subsequently penetrated and blocked with 0.1% Triton X-100 and 3% BSA for 30 min at room temperature. Then, the cells were incubated with primary antibody (H2AK119ub, CST, #8240; SS18-SSX, CST, #72364; SS18, Santa Cruz, sc-374299) for 1.5 hour. After 5 washes in PBS, one hour of incubation in secondary antibodies (Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568, A11011; Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488, A-11008; Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488, A-11001; Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647, A-21245), cells were then incubated in DAPI (Sigma-Aldrich, D9542) for 2 min. Then, the coverslips were mounted on the slides for observation on the confocal microscope (ZEISS, LSM 900).
1,6-hexadiol stimulation experiment
SS18-EGFP and SS18-SSX1-EGFP proteins were overexpressed in BJ cells, respectively, and then planted at the bottom of a glass culture dish of about 0.16 mm. Cultivate overnight in a 37 °C, 5% CO2 incubator to allow complete adhesion to the wall. Relevant experiments were filmed using a Zeiss 900. Open the temperature control module of the Zeiss 900 instrument to provide a suitable environment for the cells. Take the Zeiss 900 laser channel and set the appropriate laser intensity before starting. Carefully remove the supernatant with a pipette, quickly add the medium containing 1,6-hexadiol. Processing duration was recorded, and the condensate state was observed. Imaging was performed with Zeiss 900 confocal microscope.
Fluorescence bleaching recovery experiment
SS18-EGFP and SS18-SSX1-EGFP proteins were overexpressed in BJ cells, respectively, and then planted at the bottom of a glass culture dish of about 0.16 mm. Cultivate overnight in a 37 °C, 5% CO2 incubator to allow complete adhesion to the wall. FRAP experiments were performed using a Zeiss 900 equipped with a fluorescence bleach recovery module. Turn on the temperature control module and CO2 in advance before use to provide suitable growth conditions for living cells. Before fluorescence bleaching, take two consecutive photos to make sure that the selected shooting laser intensity will not bleach the EGFP protein. Bleaching is performed using high-energy laser photography, with a laser wavelength of 488 nm. The diameter of the bleached area is ~1 μm. After bleaching, a picture was taken approximately every 1 s until the fluorescence intensity no longer changed. After the experiment is completed, Zeiss’ own software is used to process the image and collect fluorescence intensity values. After normalization, prism software was used to perform curve fitting to draw a fluorescence bleaching recovery curve, and prism software was used to calculate the average recovery half-life value.
Detection sensitivity of condensates to high temperatures
The temperature sensitivity of phase-separated proteins can be considered as one of the characterized properties of their phase separation capabilities. We first seeded BJ cells overexpressing the SS18-EGFP and SS18-SSX1-EGFP proteins in 24-well plates lined with glass slides. Set the incubator temperature to 40 °C and 42 °C. The 24-well plates were then transferred to the corresponding incubator. Cells were fixed with 4% paraformaldehyde immediately after the appropriate duration. The state of the condensates was observed under an inverted fluorescence microscope. Cells were permeabilized with 0.1% Triton X-100 and stained with DAPI. Then, the coverslips were mounted on the slides for observation on the confocal microscope (ZEISS, LSM 900).
EdU cell proliferation assay
The proliferation of cells was detected using EdU cell proliferation assay according to the reagent instructions. About 3 × 104 cells were seeded in 48-well plates and maintained for 24 h before the assay. A total of 200 µL EdU (10 µM) reagent (Beyotime, C0078S) was added to each well and incubated for 6 h to label the cells. After two times wash with PBS, cells were fixed in a 4% paraformaldehyde solution (Sangon, E672002-0500) for 30 min, permeabilized with 0.3% Triton X-100 (Sigma-Aldrich, 9036-19-5) for 15 min, and then incubated with the click-reaction reagent for 30 min at room temperature in the dark environment. After the end of each of the above steps, wash three times with PBS containing 3% BSA for 3–5 min each time. In all, 1× Hoechst33342 reagent was used to counterstain the nucleus. The result of staining was observed with a fluorescence microscope system ZEISS, and the data were collected by the ImageJ software.
Co-immunoprecipitation experiment
First, EGFP (Control), SS18-SSX1-EGFP-Flag (SS-Flag), and SS18-SSSX1(AR Mut)-EGFP-Flag (AR Mut-Flag) proteins were overexpressed in 293T cells, respectively. Ten million cells were harvested three days later for the co-immunoprecipitation experiments. Then, resuspend cell pellet in 200 µl ice-cold lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.1% SDS, 1% Triton X-100) supplemented with protease inhibitor cocktail and 1 mM PMSF. Sonasonic with an ultrasound instrument (Bioruptor plus, SIA-UH008) for 12 cycles. Place the tube on ice for 30 min and extensively pipette the suspension every 10 min. Centrifuge cell lysate at 12,000 × g for 10 min at 4 °C. Transfer cleared lysate (supernatant) to a pre-cooled tube and add 300 µl Wash buffer(10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA) supplemented with 1 mM PMSF and protease inhibitor cocktail. 100 µl of the diluted lysate was saved for further analysis (input). Add 5x SDS-sample buffer into input sample, after 99 °C for 10 min, and set aside. After, Preparation of the beads and the protein binding. Resuspend the Flag beads (DYKDDDDK tag Nanoselector Magnetic beads, Code: 016-101-003) by gently pipetting up and down or by inverting the tube. Transfer 25 µl of bead slurry into 1.5 ml reaction tube. Add 500 µl ice-cold wash buffer. Separate the beads with a magnet until the supernatant is clear. Discard the supernatant. Add diluted lysate to the equilibrated beads. Incubate 4 °C spin for 1 h. At last, Washing and Elution with 1× SDS-sample buffer. Separate the beads with a magnet until the supernatant is clear. Discard the remaining supernatant. Washed four times with 500 µl wash buffer. During the last washing step, transfer the beads to a new tube. Remove the remaining supernatant. Resuspend beads in 100 µl 1× SDS-sample buffer. Boil beads for 10 min at 99 °C to dissociate immunocomplexes from beads. Separate the beads with a magnet. Supernatants were subjected to SDS–PAGE and incubated with corresponding primary antibody(Anti-DYKDDDDK tag, AlpHcAbs® Rabbit antibody, Code: 016-203-001; H2AK119ub, CST, #8240) and secondary antibodies(HRP-labeled Goat Anti-Rabbit IgG(H + L), Beyotime, A0208).
CUT&Tag and data analysis
The CUT&Tag experiments of SS18, SS18-SSXand H2AK119ub were constructed with NovoNGS CUT&Tag 3.0 High-Sensitivity Kit for Illumina (novoprotein N259-YH01) according to the manufacturer’s instructions. Reads form ChIP-seq experiments were mapped to the human genome (hg38) using Bowtie2 (–very-sensitive), and only those reads that mapped once were retained for further analysis. Peaks were called using MACS2 software with the default parameters. The antibodies used were SS18 (CST, #21792), SS18-SSX (CST, #72364), H2AK119ub (CST, #8240), HA (CST, #3724S), Goat anti-Rabbit IgG (Proteintech, B900210) and Rabbit anti-Mouse IgG (Proteintech, B900120).
Statistical information
Data are presented as mean±s.d. as indicated in the figure legends. Unpaired two-tailed student t test, The P value was calculated with the Prism 6 software. A P value < 0.05 was considered as statistically, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. No statistical method was used to predetermine the sample size. The experiments were not randomized. The investigators were not blinded to allocation during the experiment and outcome assessment.

Responses