Continuous time and dynamic suicide attempt risk prediction with neural ordinary differential equations

Introduction
More than 700,000 people die by suicide worldwide annually, according to the World Health Organization1. In the US, suicide rates continue to increase despite calls for and efforts in suicide prevention2. One of the cornerstones of effective suicide prevention is identifying individuals at high risk for suicide-related behaviors (SRBs) to enable early intervention. Healthcare settings provide an important opportunity for risk assessment as many people who attempt or die by suicide are seen by a healthcare provider in the preceding weeks3. In recent years, the application of statistical and machine learning models to healthcare data has shown promise in improving prediction of suicide-related behavior4,5,6,7,8,9,10. Nevertheless, whether based on clinician evaluations alone or in conjunction with newer data-driven approaches4,5, current assessment methods are largely confined to specific time points and settings, such as following a healthcare visit, and treat risk as a static estimate over a given prediction window (Fig. 1a). Such approaches are intrinsically limited given the dynamic nature of SRB risk, potentially compromising efforts to improve prevention. The precision of static risk estimates will decay with increasing intervals since the time point of prediction, progressively misaligning estimated and actual risk levels. In addition, in the absence of frequent healthcare visits, gaps in the availability of SRB risk estimates would occur, which could contain periods of high risk (i.e., the “uncovered time” depicted in Fig. 1a)). Ideally, an effective SRB risk estimate tool would provide a continuous-time, dynamic trajectory of risk that covers all future time points and can be updated with ongoing observations (Fig. 1b).
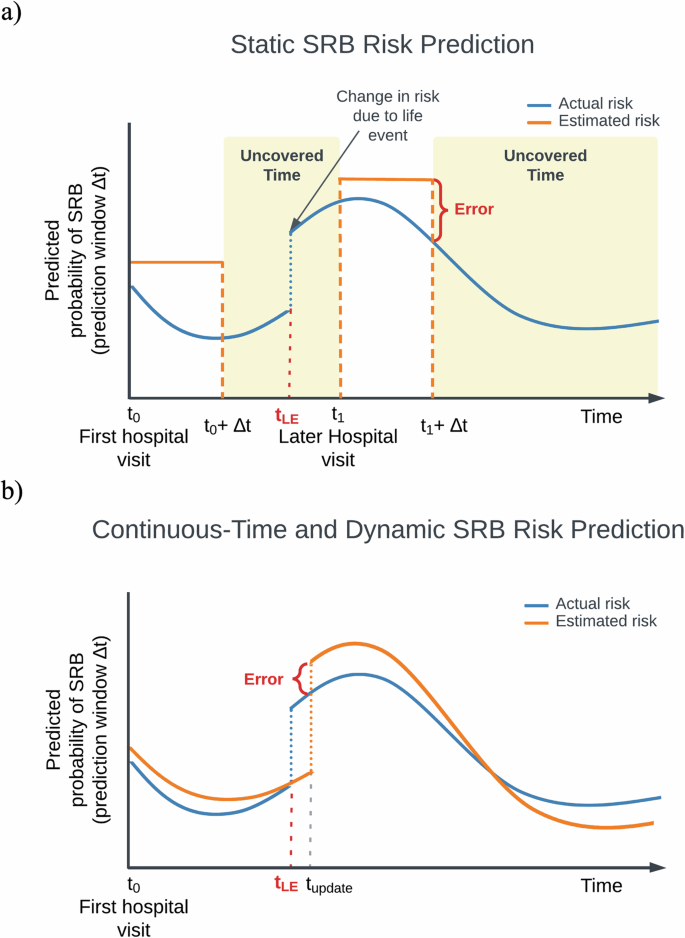
a A static paradigm. Here, risks of SRB are modeled as uniform probabilities for a pre-defined prediction window (Δt) evaluated at a specific time point, usually following a hospital visit (e.g., t0, t1). Δt may vary in length based on the query. The estimated static risk is carried over through the entire prediction window to represent the SRB risk at any given point until the end of the prediction window (e.g., between t0 and t0 + Δt). This approach is vulnerable to errors due to its inherent mismatch with the dynamic nature of SRBs, and fails to provide comprehensive coverage of estimated risk throughout the entire trajectory. On the x-axis: time since the first hospital visit; on the y-axis: actual (blue) and modeled (orange) SRB risk (cumulative incidence) for a given prediction window. t0: the time of the first hospital visit; t1: a subsequent hospital visit when new information regarding a life event is recorded; Δt1: the corresponding prediction window queried; Error: the discrepancies between estimated and actual risks; tLE: the time a life event occurred, potentially affecting SRB risk; Uncovered time: periods when an estimated suicide risk is not available; b An ideal continuous-time and dynamic SRB risk prediction paradigm. In this paradigm, risks are dynamically modeled to fluctuate naturally, i.e., for every time point, the model produces an estimated SRB risk for a given prediction window following that time point, thus reducing errors associated with static approach. The continuous-time approach also provides coverage of the entire time trajectory (i.e., no “uncovered time”). tupdate: the time when new information, such as a life event, is recorded and incorporated into the risk model. Notably, if data from sources outside healthcare settings is available, tupdate can occur independently of hospital visits, allowing for more timely updates based on the information source. Similarly, equating the timing of SRB events with the recording of diagnosis codes can introduce inaccuracies in the modeled trajectories. Further details are discussed in the “Discussion” section.
In this study, we aimed to develop and validate a first iteration of continuous-time, dynamic risk prediction models for SRBs. We utilize data from a large-scale electronic health records (EHR) database, encompassing more than 1.7 million patients with a wide range of demographic and clinical features, to employ a recent advance in artificial intelligence (AI): Neural Ordinary Differential Equations (Neural ODEs)11. Neural ODEs are a type of deep learning model that perceives the sequential evolution of data as continuous-time differential equations, allowing for more flexible and efficient modeling of complex, dynamic systems. Specifically, our models leverage GRU-ODE12, an innovation that extends the original Neural ODE framework by building on Gated Recurrent Units (GRUs)13, a specialized type of recurrent neural network. GRU-ODE is particularly well-suited for modeling the irregularly spaced and sparse longitudinal trajectories such as those captured in her data. We further adapt GRU-ODE and develop two closely related model classes for event-based predictions (e.g., a recorded SRB), which we term “Event-GRU-ODE” and “Event-GRU-Discretized.” In this framework, each patient’s SRB risk at each time step is modeled as a non-linear transformation of a latent, continuous, and dynamic trajectory. Conceptually, such a trajectory can be interpreted as a series of abstract vectors representing the evolution of mental states), enabling the estimation of time-varying probabilities of SRBs within any specified future time window. These models have an “interleaving” design – i.e., they consist of two components—one that models a base latent trajectory of risk, and another that updates the trajectory based on information acquired from new observations. Both Event-GRU-ODE and Event-GRU-Discretized are capable of producing continuous-time predictions, with the only difference being that the former imposes an additional continuity assumption on the modeled latent trajectory – i.e., in the absence of new information, the change in the modeled value of the latent trajectory cannot exceed a threshold with respect to each time interval (see Methods for details). In short, these models can estimate continuous changes in risk across the continuum of future time points, even in the absence of new observations, and can be updated when new observations become available. By extending SRB risk estimates across the continuum of time, this approach effectively addresses one of the major limitations of current SRB risk assessment methods and could serve to inform more responsive interventions that are better able to address the complex nature of suicide risk.
Results
Patient cohort characteristics
Data were obtained for 1,706,417 patients from the Research Patient Data Registry (RPDR)14, the EHR data repository of the Mass General Brigham (MGB) Healthcare system. From this patient set, 1,536,179 were randomly sampled for model development (80% of all patients for training, and 10% for validation/hyperparameter tuning), and 170,238 patients (10%) were included as a hold-out test set. Table 1 summarizes the demographic composition of the data sets. Overall, the patients were predominantly females (i.e., 58% females in both training and test sets), and a majority self-identified as White (77.8% in the training/validation sets, and 77.6% in the test set) and were in the age range of 45–65 years old (Table 1 and Supplementary Fig. 1). As shown in Supplementary Fig. 2, most patients had fewer than 50 healthcare encounters within our “study time frame” (i.e., between Jan. 1, 2016 and Dec. 31, 2019).
Our models continuously generate predictions on a daily basis for each patient, with each prediction varying in its temporal distance from both the patient’s first recorded observation and the last observation before the prediction was made. We illustrate the distribution of the number of months between the prediction time point and (1) the first observation (representing the total length of information available for the prediction in question) and (2) the previous observation in Supplementary Fig. 3. Most predictions were made with less than 2 years of data from the first observation and less than 5 months from the previous observation. See below for the effect of length of observed patient history on model performance.
Main model performance metrics
Table 2 shows the prediction performance of the Event-GRU-ODE and Event-GRU-Discretized models. In general, both models achieved excellent discrimination (area under the receiver operator curve, AUROC > 0.9) across different prediction windows (ROC curves are shown in Supplementary Fig. 4). All model metrics were reported at a fixed 95% specificity. The highest AUROCs were recorded with a 1-month prediction window (Event-GRU-ODE = 0.942, Event-GRU-Discretized = 0.941), while the highest area under the precision-recall curve (AUPRC) and positive predictive value (PPV) were observed using a 1.5-year prediction window, for both models (AUPRC = 0.095 and PPV = 0.020 for Event-GRU-ODE; AUPRC = 0.097 and PPV = 0.020 for Event-GRU-Discretized). All AUPRCs and PPVs were less than or equal to 0.1 and 0.02, respectively, due to the low SRB prevalence in the data (0.01–0.12%). Although we observed the lowest PPV with the shortest prediction window, those classified as high risk were 15 times (RR = 15.21 for Event-GRU-ODE; RR = 15.32 for Event-GRU-Discretized) more likely to have an SRB than those classified as low risk. Both models had nearly identical AUROC, AUPRC, and PPV. The Event-GRU-Discretized model achieved slightly higher sensitivity and relative risk (at 95% specificity) by point estimates with longer prediction windows (e.g., 1 year or 1.5 years).
Effect of time length of observed patient history and time since the previous observation on model performance
Figure 2a and b plot model metrics for the Event-GRU-ODE model as a function of the time length of observed patient history, smoothed by the locally weighted scatterplot smoothing (LOWESS) method and bootstrapped with 95% confidence intervals, for 1-month, 3-month, and 6-month prediction windows (1- and 1.5-year prediction windows demonstrate similar patterns; plots for the shorter prediction windows are illustrated here for simplicity). In general, despite some variances, performance tended to be better with longer observed patient history trajectories (and, therefore, more accumulated information), except for the AUPRC curve using the 1-month prediction window. We observe wider confidence intervals around the LOWESS fits towards the end of the follow-up period and with longer prediction windows. This widening may be attributed to a combination of factors: patients moving in and out of the hospital system, resulting in not all being followed for the entire study period; and the exclusion of time steps from the model training when their prediction windows extend beyond the study period. Both of these factors contribute to a reduction in the amount of data available for training during the later time steps, relative to the initial records of each patient. Using identical procedures, Fig. 2c and d plot the model metrics for the Event-GRU-ODE model as a function of the time since the previous observation. This evaluation measures the model’s capability of looking into the future without new, incoming information (i.e., using the continuous propagation component to extrapolate dynamic future risks). This capability is crucial in extending the accessibility of suicide risk estimates from specific time points into the continuum of time. Despite a minor decline in model metrics as time moves farther from the previous observation, which was expected, the models demonstrated high discriminative performance for at least one year since the last information update (AUROC ~ 0.90 across different prediction windows). The aggregated prediction plots for the discretized model are shown in Supplementary Fig. 5.

a AUROC by the number of days since the first observation; b AUPRC by the number of days since the first observation. These represent the effect of the length of observed history on prediction performance; and c AUROC by the number of days since the previous observation; d AUPRC by the number of days since the previous observation, AUPRC. These evaluate the models’ capabilities of looking into the future without new, incoming information (i.e., using the continuous propagation component to extrapolate dynamic risk into the future). The latter set of plots c, d is truncated at one year (365 days) because moving prediction windows further into the future may not be of clinical relevance, and the number of positive labels becomes too small to derive meaningful metrics. For all plots, the longer prediction windows (1 and 1.5 years) demonstrate similar trends; plots for the shorter prediction windows are illustrated here for simplicity.

Model performance for Event-GRU-ODE for three clinical settings are presented here: “General” (general setting, green), “Psych ED” (psychiatric emergency department, blue), and “Psych Inpatient” (psychiatric inpatients, orange). The percentages in the parentheses along the x-axis indicate the prevalence of SRB in each setting.
Subgroup analysis by clinical settings and demographic factors of interest
Figure 3 presents model metrics for Event-GRU-ODE stratified by three different clinical settings: (1) “General,” including all available patients and visits; (2) “Psych ED,” for which predictions were made at the time of an emergency department visits involving psychiatric evaluation/consultation; and (3) “Psych Inpatient,” for which predictions were made at time of a psychiatric inpatient admission. AUPRC and PPV were significantly higher for the “Psych ED” and “Psych Inpatient” cohort compared to the “General” cohort, presumably due to the increased prevalence of SRB events among the two sub-cohorts the across different prediction windows (2.55% to 12.63% for “Psych ED”, and 0.78% to 7.86% for “Psych Inpatient”, Supplementary Table 1). The highest PPV (0.405) was achieved for the Psych Inpatient setting, with a prediction window of 1.5 years.
Event-GRU-ODE performance metrics stratified by important patient demographic characteristics (i.e., gender, self-reported race, age, income, and public payor for healthcare) are shown in Supplementary Figs. 6–10. In general, AUROCs demonstrated modest variation across different demographic groups but were good (AUROC > 0.8) in all settings. There was greater variation in AUPRCs which were highest among individuals whose self-reported race was Black (for prediction windows of 6 months or more), and also tended to be higher among individuals aged 20–60 years and those gender was male. The discrete model version demonstrated similar characteristics in both subgroup analyses.
Effect of training data size on model performance
The size of available training data can be a limiting factor in settings with constrained resources. Theoretically, if the continuity assumption underlying the Event-GRU-ODE model holds true, this assumption should benefit model performance when training data is limited. To examine the overall effect of training data size on performance across both model versions, as well as the impact of the continuity assumption (demonstrated by contrasting the ODE and discretized models), we present in Table 3 the results (in AUROC and AUPRC) from training both models with various data sample sizes (i.e., 1/8, 3/8, 5/8, and 8/8 or 100% of the training set). Additionally, we plot the comparison of prediction performance (in AUROC, AUPRC, and PPV at 95% specificity) between the ODE and discretized models for the 3-month prediction in Fig. 4. We choose the 3-month (i.e., 90-days) prediction window due to the clinical utility of shorter-term prediction intervals. In general, model performance was only modestly reduced when training data was 3/8 (37.5%) of the full training dataset. We observe a more significant decline in model performance when the training data size is reduced from 3/8 to 1/8 of all available training data. However, discriminative metrics were still robust (AUROC > 0.88 for both model classes) under the 1/8 training data setting. Of note, Event-GRU-ODE achieved higher AUPRC (a model metric which have been shown to be more informative than AUROC when base prevalence is low15,16) compared to Event-GRU-Discretized with smaller training data sizes, i.e., with only 5/8, 3/8 or 1/8 of training data. Performance plots for the other prediction windows are shown in Supplementary Figs. 11–14.

For each panel, a model metric (with a 3-month prediction window) is plotted versus the amount of training data used (1 fold, 3 folds, 5 folds, and 8 folds; i.e., 1/8, 3/8, 5/8, and the entire training set): a AUROC; b AUPRC; and c PPV at 95% specificity.
Predictor (feature) importance
The top ten predictors for the Event-GRU-ODE model across all prediction windows, identified through a predictor ablation method (see Methods) and evaluated using AUROC, AUPRC, and PPV, are detailed in Supplementary Table 2. For NLP features, the change estimate represents the mean of change associated with positive mentions, negated mentions, and mentions or negations of family history (see Methods). The top predictors are primarily associated with psychiatric history, age, and mentions of behavioral factors (e.g., exercise) and past treatments (e.g., electroconvulsive therapy) in clinical notes (derived from extracted “NLP CUIs”; see “Methods”). Notably, the absolute contribution of each feature—and the differences between features—are modest. For example, the average contribution of the top 100 features to AUROC (0.009) is close to the contributions of the top 10 predictors. This likely reflects the inclusion of a large number of predictors (>6000) and underscores the understanding that SRBs arise from complex interactions among numerous factors, rather than being driven by a few dominant contributors. Finally, we note that feature importance varies depending on the performance metric of interest, making interpretation challenging. This is not surprising given that this method of extracting feature importance, like many others, depends on the complex correlational structure among features and how this structure interacts with the model.
Discussion
In this study, we present a continuous-time, dynamic modeling approach for predicting SRB risks. Unlike current assessment methods, this approach accommodates the time-varying nature of SRB risks and, by generating risk estimates across the entire time trajectory, it mitigates gaps in risk score availability. We utilized data from a large-scale EHR database containing data from multiple medical centers and hospitals, including features extracted from unstructured clinical notes, to develop and validate two model classes (Event-GRU-ODE and Event-GRU-Discretized) based on an advanced neural network architecture—Neural ODEs. Both model classes achieved high discrimination performance across different clinical settings. For example, in the general setting, AUROCs were between 0.93–0.94, and relative risks (the modeled risk divided by baseline prevalence during the designated prediction window) ranged from 13.4 and 15.3 across different prediction windows at 95% specificity. Such performance is better or comparable to those observed in previous analyses based on conventional static risk estimates by our group and others4,6,7,8,9,10,17,18,19.
With respect to analyses by clinical setting, the results largely tracked the base rate of SRBs. As expected, AUROCs were generally higher and PPVs lower for the “General” (all patients) cohort than the inpatient or ED sub-cohorts where base rates are higher. Stratified analyses by demographics found good discrimination across groups (AUROCs > 0.80) while AUPRCs were more variable (again, consistent with varying base rates of the outcome). For prediction windows of 6 months or longer, AUPRCs tended to be highest among those who identified as Black and lowest among those who identified as Asian, for those with their race reported.
While the two model classes achieved similar performance across various metrics when all training data were used, Event-GRU-ODE attained higher AUPRC than its discretized counterpart for the majority of configurations in the settings where the size of training data is limited. This implies that the continuity prior assumption12 (see Methods) may be beneficial in such settings.
While these proof-of-concept analyses focused solely on EHR data for model development, a key advantage of our model classes lies in their flexible architecture, including their natural incorporation of a time dimension. This flexibility facilitates the easy integration of additional data modalities, such as genetic data or other biomarkers, and enables updates to the modeled future risk trajectory based on new incoming information at any point in time. This is crucial because, although a risk trajectory generated at a certain point can theoretically cover all future times, it may diverge from reality if it extends too far into the future without incorporating updated information. Sources of new information can include future healthcare visits, data from digital sensors, or outputs from mobile devices, which may themselves be structured as time series. Crucially, updating the modeled risk trajectory with incoming information at a specific time step requires only the modeled latent state vector from the previous time step, the relevant new information content, and the model parameters. This approach eliminates the need for access to the original data utilized to generate the risk trajectory up to the current time point, which may contain protected health information, thereby mitigating privacy concerns when updating predictions outside healthcare facilities.
Our results should be interpreted with several limitations in mind. First, in this initial iteration of continuous-time SRB prediction models, our models rely solely on structured EHR data and natural language processing (NLP)-derived concepts from clinical notes. Importantly, the timing of medical or related events, including SRBs, may be imprecise, as it often reflects when structured codes are entered into the system. This can lead to delays for past events (e.g., an SRB event) or premature timing for planned events (e.g., taking prescribed treatments) relative to their actual occurrence (Supplementary Fig. 15). A natural next step would be to refine the precision of event timing allowing for more timely updates in the model, potentially through: (1) leveraging more advanced NLP methods (e.g., large language models20,21,22) that could more accurately capture events and their timing; and (2) incorporating additional real-time or near-real-time data sources e.g., from mobile sensors or digital health applications.
Second, using ICD codes as a surrogate for SRBs is not without limitations. However, the ICD codes used to define SRB in this study have been validated through extensive chart reviews from the same EHR repository (see Methods). Follow-up studies can address this limitation by using advanced NLP techniques on clinical notes to enhance SRB phenotyping and further align with expert chart reviews. Third, model interpretation in time series modeling is an evolving field, and the interleaved design of Event-GRU-ODE and Event-GRU-Discretized—combining discrete updates with continuous propagation—presents unique interpretability challenges. Standard methods, such as SHAP (Shapley Additive Explanations)23, developed for static or traditional time-series models, are not well-suited for this hybrid architecture. In addition, the large number of time-series predictors and the size of the patient cohort pose computational challenges for exhaustive methods like feature permutation. To address these, we implemented a model explanation method based on predictor ablation on a random sample of test set patients, enabling a feasible estimation of global per-feature importance at the input level. Many of the top predictors (e.g., past psychiatric history and electroconvulsive therapy, a second-line psychiatric treatment for severe or treatment-resistant conditions) are related to known risk factors for suicide attempts. However, while the ablation results enhance transparency of the model decisions, they do not necessarily identify actionable targets for intervention. This is because supervised learning algorithms optimize performance by capitalizing on the network of correlational relationships among features so that feature importance for prediction does not necessarily reflect causal importance for the outcome. Research to address this gap between the predictive and mechanistic interpretation of features includes evolving methods to integrate prediction and causal inference (i.e., “causal machine learning”24), and future work in this area may inform the identification of intervention targets. Lastly, this initial study on continuous-time SRB prediction models has not yet explored their generalizability in depth.
Effective suicide prevention strategies necessitate a collaborative approach between the healthcare system and the broader community, including schools, workplaces, and families. The long-term goal of developing risk prediction tools that can adapt to dynamic changes in SRB risk, integrate new information continuously, and offer timely assessments, is to enhance the design of suicide prevention strategies. For example, data from community settings could be integrated into real-time risk assessments for more timely delivery of support or interventions. Alternatively, healthcare visits might be strategically scheduled during periods of predicted spikes in risk, facilitating better planning around times of the year or following specific life events that may elevate risk.
In conclusion, leveraging neural ODE—a recent advance in deep learning for time-series analysis—we have developed continuous-time and dynamic risk prediction models to assess the risk of SRB using EHR data. We validated their effectiveness across various prediction windows, with robust performance across diverse demographics and clinical settings. Our results confirm the feasibility of continuous-time and dynamic prediction for SRB and highlight the value of continued innovation in this area to enable more accurate and comprehensive SRB risk stratification methods.
Methods
Institutional review board approval
All procedures were approved by the Institutional Review Board of Mass General Brigham (MGB) Healthcare System (Boston, Massachusetts, USA, Protocol 2020P000777), with a waiver of consent granted for the analysis of electronic health record data. This waiver was justified because the retrospective analysis of clinical data conducted in this study involved no more than minimal risk to patients, did not adversely affect their rights or welfare, and obtaining consent was infeasible due to the scale of the study.
Data source and study population
The data used for this study were derived from RPDR14, the research EHR data registry of the MGB healthcare system located in Massachusetts, USA. The RPDR is a centralized data registry that collects and compiles clinical information from multiple EHR systems within MGB, which includes more than 7 million patients with over 3 billion records seen across more than 8 hospitals, including two major teaching hospitals14. In this study, we used data between the period of Jan. 1, 2016, and Dec. 31, 2019 (the “study time frame”) and included all patients who had at least 3 visits and a minimum of 30 days of medical record and were older than age 10 at the time of their first medical record entry within this time period. We chose the start date to minimize the impact of data heterogeneity caused by different recording practices due to the hospital system’s transition from ICD (International Classification of Disease)-9 to ICD-10.
Suicide-related behavior definition
For the purpose of this study, we adopt a list of ICD codes, previously developed and validated8,25, as the definition of SRB. These codes were shown to be valid for capturing intentional self-harm through extensive chart review by expert clinicians, with a PPV of greater than 70% for ICD-9 codes and greater than 80% for ICD-10 codes25. Of note, because the study period began in 2016, almost all of the ICD codes used for outcomes were from the higher ICD-10 group. An “SRB event” is then defined by the occurrence of any of the codes in the list.
Predictors
We include two types of predictors: (1) covariates, including the demographic variables (total of 7, a full list is provided in Table 1). These enter the model at the beginning of the trajectories; and (2) observations, which are recorded for each hospital encounter during the patient trajectory, including a. diagnosis codes, which were mapped from ICD codes26,27 to PheWAS codes28 which have been shown to capture clinically meaningful concepts29 (total 1,871 features); b. medications, which were mapped to RXNORM codes30 (total 1712 features); and c. a set of previously defined features derived using NLP9, which we named as “NLP CUIs” (i.e., concept unique identifiers31), indicating the occurrences of mental health-related concepts (positive mentions, negated mentions, and mentions or negations of family history) as defined in the Unified Medical Language System (UMLS31) in the clinical notes (total of 2488 features). Observations were ordered temporally and entered the model as inputs according to their recorded date relative to the date of the first entry of each patient (see Fig. 5 for a schematic illustration of the data setup). Note that while we chose a time resolution of one day as the basic unit for a time step, any time resolution can theoretically be chosen given that data are available (the smaller the time step, the closer the approximation to continuity). Continuous-valued features (e.g., age range at the time follow-up started) were discretized by binning. The NLP features were represented as occurrence counts at each time step (most of which were zero or one), while all other features were transformed using one-hot encoding before being input into the models.

Static covariates, such as demographic characteristics, along with additional diagnoses, medications, and NLP CUIs, are entered into the model at time t0 (i.e., the time of the first patient entry). Subsequent observations are ordered sequentially over time, with the exact time steps (days after the first record) preserved. Labels are created for each time step, considering the presence of an SRB record. For any time step tA within the interval where data is available, its associated prediction Window A, starting from tA + 1, is assigned a label value of 1 if SRB is recorded within that period. Note that tA is not bound to the dates of where there are recorded entries. With the trained model, predictions can be made for any future time step (tB, tC, …) occurring after the last recorded entry at tf (note that labels are not available for future times and the probability of their occurrences are the targets for predictions (red circle with question mark)).
Prediction task definition
Based on the SRB definition, we formulate the suicide risk prediction task as follows:
where (Delta t) is the length of the “prediction window” and ({e}_{tau }) is 1 if an event (i.e., SRB) was recorded at time (tau). This means (Y(t)) (i.e., the prediction label) is set to 1 if an event happens within the next (Delta t) time (i.e., a “prediction window”). In this study, we look at five prediction windows of different lengths: 1 month, 3 months, 6 months, 1 year, and 1.5 years.
Prediction labels are created for each patient, for each day, and for each prediction window from the date of the first visit up to the date of the last visit, minus the length of the prediction window, less thirty days of “buffering time,” to (1) ensure the full length of prediction window is observed; and (2) to compensate for the fact that events recorded toward the end of patient history would contribute fewer labels under this scheme.
Event-GRU-ODE model
We model the occurrence of events of interest ({boldsymbol{Y}}(t)) (e.g., an SRB event, or any other event of interest) as a continuous-time stochastic process, where ({boldsymbol{Y}}(t)) is a D-dimensional random variable, with each dimension (din {mathrm{1,2},ldots ,D)} being governed by a Bernoulli distribution, i.e., ({Y}_{d}left(tright)in {0,,1}) and parameterized by probability ({p}_{d}(t),in [0,1]). However, the EHR records in a patient trajectory typically consist of sparse and irregularly observed longitudinal data, which makes directly modeling the stochastic process directly very difficult. We follow the idea from statistical mechanics, that instead of modeling the stochastic process directly, one models the time evolution of the probability mass function(,{boldsymbol{p}}left(tright)) of the observables ({boldsymbol{Y}}(t)). Therefore, we propose to use a latent variable process ({boldsymbol{h}}left(tright)) that can be mapped to the SRB labels ({boldsymbol{Y}}(t)) (see “Prediction task definition”) through their generating probability mass functions. In detail, we model the probabilities of an SRB event happening between time periods (t) and (t+Delta t) ((Delta t) indicating the prediction window) in the form of Bernoulli distributions. In this specific case, the number of dimensions of interest D = 5, with each dimension representing the trajectory of SRBs under a specific prediction window (five total) as a function of time. This means that the latent process ({boldsymbol{h}}left(tright)) is mapped to the parameters ({boldsymbol{p}}left(tright)) of the Bernoulli distributions, which determines its mean and variance varying in time. The dynamics of the underlying latent process are governed by ODEs, whose parameters are learned from data.
Specifically, we model the mapping from the observed variables to the hidden variable with “Event-GRU-ODE” as described below, and use a feed-forward neural network to map the hidden variable ({boldsymbol{h}}left(tright)) to the output probabilities (i.e., the parameters ({boldsymbol{p}}(t)) of the Bernoulli distributions). This formulation reflects the evolving uncertainty of the patient’s future condition regarding SRB.
To model the latent variable ({boldsymbol{h}}left(tright)), used for predicting the probabilities of SRB events, and incorporate incoming health-related observations, such as diagnosis and measurements, we propose a two-mode system, consisting of: (1) Continuous time propagation of the latent variable ({boldsymbol{h}}left(tright)); and (2) Discrete updates that happen each time a new observation ({{boldsymbol{x}}}_{t}) is received at time (t). Each time there is an observation that triggers the discrete update the system switches to a new state of the ODE (which still has the same parameters and dynamics as before).
For the continuous time propagation, we employ recently proposed methods in the field of Neural ODEs, specifically GRU-ODE12. This means when there are no observations, the latent variable follows an ODE whose trajectory is controlled by the GRU-ODE:
where ({W}_{c}) are the weights of the neural networks of GRU-ODE (learned end-to-end with other components of the model).
To derive GRU-ODE we start with the standard classic GRU13, given by its equations for update gate ({boldsymbol{z}}), reset gate r and candidate hidden vector (hat{{boldsymbol{h}}}):
The key idea is to observe that the last equation, which combines the candidate hidden vector (widehat{{h}_{t}}) and the previously hidden vector ({h}_{t-1}), can be rewritten as a difference equation for ({h}_{t}):
Taken to the limit of (Delta {t}to 0) the difference equation gives an ODE with negative feedback on the current hidden state ({boldsymbol{h}}left(tright)):
GRU-ODE has several useful properties, such as stability due to negative feedback and Lipschitz continuity with respect to time with constant K = 2, which helps make the model robust to numerical errors as well as effective even with smaller data sample size12.
The initial hidden state, ({boldsymbol{h}}left(0right)), is computed from the static covariates of the patient using a multilayer feed-forward neural network. The continuous-time GRU-ODE component propagates the latent vector up to the observation time (left(tright)) where the model then updates the hidden state ({boldsymbol{h}}left(tright)) to a new value ({{boldsymbol{h}}}^{{prime} }) by using a discrete network, which is a standard GRU13:
where ({{boldsymbol{x}}}_{t}) is the observation at time (t) and ({W}_{d}) are the weights of the update GRU network and a multilayer feed-forward network that preprocesses ({{boldsymbol{x}}}_{t}). After this update, the system switches back to continuous-time propagation (GRU-ODE) starting from the state ({{boldsymbol{h}}}^{{boldsymbol{{prime} }}}left(tright)).
Finally, we use an output network (fleft({boldsymbol{h}}left(tright);{W}_{!o}right)) that maps the hidden state ({boldsymbol{h}}left(tright)) to the SRB event probabilities (p(t)) for the time (t). Using cross-entropy loss on the outputs of the network and feeding in observations ({{boldsymbol{x}}}_{t}) we can train Event-GRU-ODE in fully end-to-end manner.
Event-GRU-Discretized model
The Event-GRU-ODE model makes the assumption that between two observations (e.g., two healthcare encounters) the latent vector ({boldsymbol{h}}) is propagating by following an ODE, forcing the trajectory to be continuous in time.
We can relax this continuity assumption by allowing the time propagation to be updated by the discrete GRU. This propagation is still carried out in iterative fashion using time step size as in Event-GRU-ODE. Relaxing the continuity assumption could allow the model to be more flexible and might affect performance. We name this model “Event-GRU-Discretized.”
Model training and evaluation
We implemented and evaluated two models, namely “Event-GRU-ODE” and “Event-GRU-Discretized”, for the continuous time suicide risk prediction task, with 5 different prediction windows (i.e., 1 month, 3 months, 6 months, 1 year, and 1.5 year). We randomly split the data into 10 “folds,” each containing data from 10% of all patients. Among them, 8 folds (80%) were used as a training set, one fold (10%) as a hold-out validation set (10%) for tuning all the model hyperparameters, and one fold (10%) as a hold-out test set for model evaluation (10%). During training, models were optimized using the Adam optimizer32. More detailed training procedures, including hyperparameter settings, are provided in Supplementary Note no. 1.
For model performance evaluation, we report AUROC, AUPRC, as well as PPV and sensitivity with specificity set to 0.95.
In order to evaluate how well Event-GRU-ODE makes its predictions as a function of the length of observed patient history trajectories, we aggregated all the model predictions made on each day after the first EHR entry of each patient, measured model performance (in AUROC and AUPRC) along the time trajectory, and plotted the model metrics over time for the study time frame. To increase the signal-to-noise ratio, plots were smoothed by the LOWESS method with its smoothing window (i.e., the fraction parameter) set to be 0.3. We also performed bootstrap resampling to add the estimated 95% confidence intervals around each LOWESS fit on the plot. Furthermore, to evaluate the models’ capabilities of looking into the future to predict continuous, dynamic risk trajectories without incoming new information (i.e., using the continuous propagation component to extrapolate time-varying future risks), we performed the same plotting procedure as above, but changing the x-axis to the number of days since the last observation.
In addition, we assessed potential differences in model performance across various patient subgroups. First, we defined three patient groups of interest based on clinical settings to stratify prediction results and evaluate model performance in each patient population: (1) “General” representing the general outpatient cohort, which encompasses the full study population and their complete trajectories within the study period; (2) “Psych ED” for predictions during emergency department visits involving psychiatric evaluation/consultation within the study period; (3) “Psych Inpatient” for predictions made during psychiatric inpatient admissions within the study period. Furthermore, we conducted stratified analyses on key demographic groups (gender, race, age group, income level by ZIP code, whether the patient has a public insurance payor) to examine potential heterogeneity in model performance across different subgroups.
Lastly, to further compare the two models and their capabilities to learn from fewer samples, we trained each model using all the training data as well as randomly sampling 5/8, 3/8 and 1/8 (i.e., 5, 3, and one fold, respectively) of the training data.
Model interpretation
To evaluate how Event-GRU-ODE weights the contribution of each predictor, we developed a feature importance ranking algorithm based on predictor ablation. Using a 20% random sample from the general cohort test set, we assessed importance by measuring changes in model performance, evaluated via AUROC, AUPRC, and PPV, across prediction windows when individual predictors, such as diagnosis codes, were excluded using a binary mask. The mask set the predictor of interest to zero, effectively removing it from the input. The ablation process involved systematically excluding all observations related to a given predictor, comparing results to those generated with the full predictor set, and repeating this for each predictor. This approach was chosen for its simplicity and model-agnostic design, which is ideal for our model’s complex, interleaving structure.

Responses