COVID-19 health data prediction: a critical evaluation of CNN-based approaches

Introduction
The COVID-19 pandemic has presented unparalleled challenges to global public health systems, prompting the urgent need for advanced and reliable prediction models to support disease management, policy formulation, and containment strategies1,2,3,4. Such models are crucial for tracking disease progression, assessing risk factors, and optimizing resource allocation in real-time. Convolutional Neural Networks (CNNs), a powerful class of deep learning algorithms, have emerged as a promising technology in this regard, demonstrating the ability to process and analyze diverse health data types, including medical images, genomic data, and time-series information. Their application in predicting COVID-19-related outcomes has garnered attention due to their potential to uncover complex patterns and correlations in high-dimensional datasets.Despite their growing popularity, the deployment of CNNs in healthcare settings, particularly for COVID-19 prediction, is fraught with challenges that must be addressed to ensure their accuracy, robustness, and practical utility. This research paper takes an exhaustive data science perspective to investigate the critical limitations of CNN-based prediction models for COVID-19 health data. The study identifies three primary areas of concern: data quality, model architecture, and generalization capabilities, each of which plays a pivotal role in shaping the performance and reliability of CNN models5,6,7.
First, data quality issues represent a significant hurdle. COVID-19 health datasets often suffer from incomplete, noisy, or imbalanced data, which can skew model training and lead to biased predictions8,9. The lack of standardized and representative datasets across different regions and populations further complicates the development of generalizable models. This paper highlights the need for enhanced data curation practices, including robust preprocessing techniques, data augmentation strategies, and the incorporation of synthetic data to mitigate these challenges. Second, architectural limitations of CNNs are explored, with particular emphasis on their reliance on extensive computational resources and sensitivity to hyperparameter settings. The paper discusses how these constraints can hinder the scalability and efficiency of CNNs, especially in resource-constrained settings. Moreover, the complexity of CNN architectures can lead to overfitting when trained on limited datasets, reducing their ability to perform reliably on unseen data10,11,12,13,14. Strategies such as transfer learning with pre-trained networks like ResNet and EfficientNet, as well as the integration of advanced optimization techniques, are proposed to address these issues. Third, generalization remains a critical concern for CNN models in the context of COVID-19. Models trained on specific datasets often struggle to adapt to variations in data distribution across diverse populations and clinical environments15,16,17. This lack of adaptability undermines the credibility of predictions in real-world applications. The paper underscores the importance of cross-validation, external validation on independent datasets, and the adoption of multimodal approaches that incorporate complementary data sources, such as patient demographics and laboratory results, to enhance the robustness of predictions.
By systematically elucidating these challenges, this research provides actionable insights to researchers and practitioners aiming to deploy CNNs for COVID-19 health data prediction. The study also highlights the transformative potential of CNNs when combined with advanced methodologies, such as regularization techniques, focal loss functions, and domain-specific adaptations, to overcome existing barriers. Ultimately, this paper contributes to the broader discourse on the role of deep learning in healthcare, offering a roadmap for the development of robust, scalable, and clinically relevant CNN-based models. By addressing the limitations outlined, this research aims to bridge the gap between theoretical advancements and practical implementation, fostering the creation of AI-powered tools that can significantly improve public health outcomes during the COVID-19 pandemic and beyond.
Background
The COVID-19 pandemic has catalyzed a remarkable increase in the volume and diversity of health data collected across the globe. This data encompasses a broad spectrum of sources, including medical imaging modalities like chest X-rays and computed tomography (CT) scans, electronic health records (EHRs) detailing patient clinical histories, molecular profiles such as genomic sequences and protein markers, as well as time-series data from wearable health devices and continuous monitoring systems. These multifaceted datasets offer an unprecedented opportunity to leverage advanced prediction models for diagnosing COVID-19 cases, forecasting disease trajectories, predicting patient outcomes, and optimizing healthcare resource allocation. However, the analysis of COVID-19 health data introduces unique challenges that distinguish it from traditional medical datasets. During the early phases of the pandemic, the availability of COVID-19 data was constrained by several factors. First, the concentrated impact of the disease on specific regions and populations led to highly imbalanced datasets, with over-representation of certain demographic groups and under-representation of others. This lack of diversity in data introduced biases in model training and hindered the development of generalized prediction algorithms. Furthermore, the limited volume of early COVID-19 data made it difficult to train robust models capable of capturing complex patterns and correlations.
As the pandemic evolved, new challenges emerged with the appearance of novel SARS-CoV-2 variants, each exhibiting distinct characteristics in transmissibility, severity, and immune evasion. This dynamic nature of the virus required predictive models to adapt continuously, accounting for the changing landscape of clinical and epidemiological data. Models that performed well during the initial waves of the pandemic often struggled to maintain their accuracy and reliability in the face of new variants and shifting patient demographics. Such challenges underscored the importance of designing flexible and adaptable prediction frameworks that could evolve alongside the pandemic. Additionally, the integration of heterogeneous data types-ranging from imaging data to clinical and molecular information-posed technical complexities in terms of data preprocessing, standardization, and feature extraction. The lack of standardized protocols for data sharing and annotation further compounded these issues, limiting the scalability and reproducibility of predictive models across different healthcare settings. Despite these challenges, the development of effective and reliable prediction models remains a critical priority for the data science community. Predictive algorithms hold immense potential to transform the fight against the pandemic by enabling early diagnosis, risk stratification, and resource optimization. For instance, accurate prediction models can assist in identifying high-risk patients who require immediate medical intervention, forecasting regional surges in case numbers to guide public health policies, and prioritizing vaccine distribution in underserved populations.
The urgency of these needs has driven significant advancements in the application of machine learning and deep learning techniques to COVID-19 health data. CNNs, in particular, have shown promise in processing and analyzing high-dimensional data such as medical images, enabling automated and accurate predictions. However, their application has also exposed critical limitations, including sensitivity to data quality, overfitting on limited datasets, and challenges in generalizing to unseen populations. In conclusion, the COVID-19 pandemic has created an unprecedented demand for innovative and adaptive prediction models. The development of such models necessitates overcoming significant obstacles, including data imbalance, the emergence of new virus variants, and the integration of heterogeneous data sources. Addressing these challenges is vital to harness the full potential of data-driven approaches in managing the pandemic and shaping future responses to global health crises. This article explores these challenges in-depth and discusses the role of CNNs in navigating the complexities of COVID-19 health data prediction.
Motivation
Machine Learning (ML) and Deep Learning (DL), integral branches of Artificial Intelligence (AI), employ neural networks to process vast datasets and uncover complex patterns. Their adaptability has driven transformative advancements across numerous domains18,19,20,21,22,23,24,25,26,27,28,29. In the healthcare sector, deep learning has become indispensable in areas such as medical imaging, drug development, predictive analytics, personalized treatment strategies, and robotic-assisted surgery30,31. By delivering highly accurate and efficient solutions, deep learning continues to reshape industries and enhance quality of life. However, challenges such as ensuring data privacy and addressing significant computational resource requirements remain critical for its sustainable advancement.
Objectives of the study
The primary objective of this research is to conduct a comprehensive investigation into the challenges and limitations associated with the utilization of CNNs for COVID-19 health data prediction. The study entails a meticulous exploration of the complexities related to data collection, preprocessing, and model architecture, with the overarching aim of identifying potential factors that may negatively impact the accuracy and generalizability of CNN-based predictive models. Additionally, the research endeavors to provide real-life case examples that exemplify the practical implications of these challenges, while proposing feasible mitigation strategies to enhance the robustness and reliability of CNN-based prediction models in the context of COVID-19 health data. This scholarly inquiry aims to contribute valuable insights to the data science community and offer support to ongoing endeavors in the effective management and control of the COVID-19 pandemic.
Scope and limitations
This study is dedicated to investigating the challenges and limitations concerning the application of CNN in the domain of COVID-19 health data prediction. The research specifically focuses on examining the diverse array of health data sources used in this context, encompassing radiological images, clinical records, and genetic profiles. By conducting a thorough analysis, the paper aims to furnish a comprehensive overview of the impediments encountered while employing CNNs for predictive modeling with these varied data sources. Moreover, this research endeavors to provide valuable insights into potential enhancements that can be made to develop more dependable and interpretable predictive models. Key factors considered include data scarcity, inherent biases, computational complexities, and the ability of the models to generalize effectively to novel COVID-19 variants. Real-life examples and empirical analyses are presented throughout to support the findings, and the paper concludes by offering valuable recommendations for researchers and practitioners in the data science field, making significant contributions to the global endeavors in managing and mitigating the COVID-19 pandemic32,33.
Literature review
COVID-19 data prediction techniques
The outbreak of the COVID-19 pandemic, caused by the novel coronavirus SARS-CoV-2, has had far-reaching global implications on both public health and the economy. The rapid transmission of the virus and its continuous mutation into new variants have put immense strain on healthcare systems worldwide, necessitating the development of effective prediction models to support disease management and control efforts. In response, data science researchers have turned to sophisticated machine learning techniques, such as CNN to analyze vast quantities of COVID-19 health data34,35 and forecast outcomes like infection rates, disease severity, and mortality. CNNs, renowned for their ability to extract meaningful patterns from complex data, have shown promising outcomes in diverse domains, rendering them attractive options for COVID-19 data analysis. Nevertheless, it is of utmost importance to undertake a comprehensive investigation into the challenges and limitations associated with utilizing CNNs for COVID-19 health data36,37 prediction, ensuring the dependability and applicability of these models in practical scenarios. Gaining insights into these limitations is crucial in the quest to build robust and accurate prediction models that can provide valuable guidance to policymakers, healthcare professionals, and researchers, enabling them to make well-informed decisions in effectively combating the COVID-19 pandemic. This research paper aims to explore the data science perspective of utilizing CNNs for COVID-19 health data prediction, shedding light on potential obstacles that researchers may encounter and proposing plausible mitigation strategies to enhance the models’ performance and interpretability38,39.
Convolutional neural networks in health data analysis
CNNs have emerged as a groundbreaking advancement in the field of health data analysis, showcasing remarkable capabilities in various medical applications. In the context of COVID-19 health data prediction, CNNs have garnered significant attention due to their inherent ability to automatically learn hierarchical features from raw data sources, such as chest X-rays, CT scans, and molecular sequences. This attribute enables CNNs to excel in capturing spatial relationships and identifying intricate patterns, rendering them well-suited for critical tasks like COVID-19 diagnosis, severity prediction, and patient outcome prognosis. Consequently, CNNs have shown promise in differentiating COVID-19 from other respiratory diseases, thereby supporting healthcare practitioners in making timely and accurate decisions. The capacity of CNNs to extract informative features without requiring explicit feature engineering has significantly contributed to elevating the predictive performance of COVID-19-related health data analysis, prompting substantial research and development efforts40,41.
However, the implementation of CNNs in health data analysis, specifically for COVID-19, is not without challenges and limitations. A notable obstacle lies in the considerable amount of labeled data necessary for effective training, which may be scarce and imbalanced in the case of COVID-19 datasets. Additionally, the interpretability of CNNs poses another significant challenge as their black-box nature restricts researchers and healthcare providers from comprehending the underlying decision-making process. Furthermore, the potential lack of generalization to new COVID-19 variants necessitates regular model updates and retraining efforts. Despite these limitations, there are potential avenues to address these challenges through the adoption of data augmentation techniques, transfer learning methods, and interpretability approaches. By effectively addressing these issues, the utility and reliability of CNNs in COVID-19 health data prediction can be significantly enhanced, thereby contributing to improved disease management and control42.
Previous studies on CNNs for COVID-19 health data prediction
Several prior investigations have demonstrated the efficacy of CNN in the realm of COVID-19 health data prediction. For example, Wang et al. (2020) conducted a study where they applied a CNN-based model to assess the severity of COVID-19 cases based on chest X-ray images. The outcomes indicated promising results, as the model exhibited accurate classification of patients into distinct severity categories, enabling early identification and prioritization of critical cases. However, it is worth noting that the study also highlighted the challenge of data scarcity in this domain. Obtaining a large and diverse dataset of COVID-19 chest X-rays proved difficult due to concerns about patient privacy and the limited availability of positive cases during the early stages of the pandemic.
Another research endeavor undertaken by Zhang et al. (2021) involved the utilization of CNNs to predict COVID-19 mortality risk, leveraging electronic health records (EHRs) from a comprehensive hospital database43. The study revealed that CNNs effectively extracted pertinent patterns and features from the EHRs, resulting in accurate mortality risk predictions. Nonetheless, a notable limitation of this approach pertained to the potential bias present in the EHR data, leading to potential disparities in predictions for specific patient cohorts. Furthermore, the CNN model’s lack of interpretability raised concerns among healthcare professionals, as comprehending the decision-making process was imperative for establishing trust and facilitating clinical adoption. These studies underscore the significance of addressing data-related challenges and limitations when deploying CNNs for COVID-19 health data prediction.
Methodology
Data collection and preprocessing
In this study, we collected COVID-19 health data from diverse publicly accessible datasets, including the COVID-19 Image Data Collection, COVID-19 Open Research Dataset (CORD-19), and electronic health records from various hospitals. The dataset encompassed chest X-rays, CT scans, and clinical information of patients diagnosed with COVID-19. To ensure the integrity and reliability of the data, extensive preprocessing measures were undertaken, encompassing data cleansing, image normalization, and addressing missing values. To address the potential imbalance between COVID-19 positive and negative cases, we implemented oversampling and data augmentation techniques to achieve a balanced dataset, thus mitigating any inherent bias in the model. A representative subset of 10,000 images, maintaining an equal distribution of COVID-19 positive and negative cases, was utilized for the purpose of training and validation.
CNN model architecture
In response to the challenges and limitations inherent in COVID-19 health data prediction, we developed a specialized CNN model, specifically tailored for medical imaging analysis. The architecture of the model was carefully designed to address the intricacies of COVID-19 data and improve prediction accuracy. It consisted of multiple convolutional layers, followed by batch normalization, max-pooling, and dropout layers to mitigate potential issues of overfitting. To harness the power of transfer learning and leverage existing knowledge from pre-trained CNN models, such as VGG-16 and ResNet, we integrated these models to extract meaningful features from COVID-19 images. The final layers of the CNN were customized for binary classification, distinguishing between COVID-19 positive and negative cases.
For implementation, we utilized TensorFlow and Keras as the framework, optimizing the model with the Adam optimizer and employing binary cross-entropy as the loss function. To assess the model’s ability to generalize to new data, a 5-fold cross-validation approach was employed, ensuring robust evaluation. Model performance was evaluated using multiple metrics, including accuracy, precision, recall, F1-score, and the area under the receiver operating characteristic curve (AUC-ROC). This comprehensive evaluation provided a holistic view of the model’s predictive capabilities, ensuring its effectiveness and reliability in handling COVID-19 health data prediction tasks.
Evaluation metrics
To evaluate the efficacy of the CNN model in predicting COVID-19 health data, we employed a set of rigorous evaluation metrics. The accuracy metric was utilized to assess the overall correctness of the model’s predictions, providing a measure of its ability to correctly classify cases. Precision and recall were utilized to gain insights into the model’s capacity to accurately identify COVID-19 positive cases and minimize false positives. The F1-score was adopted as a balanced metric, considering both precision and recall to ensure a comprehensive evaluation of the model’s performance. Moreover, the Area Under the Receiver Operating Characteristic Curve (AUC-ROC) was employed to gauge the model’s ability to distinguish between positive and negative cases at different probability thresholds.
To address potential issues of overfitting, we monitored the training and validation loss throughout the training process. Early stopping was implemented as a precautionary measure, terminating the training if the model failed to exhibit improvement on the validation set. By comparing the final model’s performance with baseline models and state-of-the-art approaches, we were able to gauge its effectiveness in overcoming the challenges and limitations inherent in COVID-19 health data prediction. The careful selection of evaluation metrics and monitoring procedures allowed for a thorough assessment of the model’s predictive capabilities and its potential utility in real-world scenarios.
In our study, we applied a systematic methodology to predict COVID-19 infections using chest X-ray images. The dataset comprised 10,000 X-ray images from diverse sources, including the COVID-19 Image Data Collection and hospitals’ radiology departments. Prior to model development, rigorous preprocessing was conducted, encompassing image resizing to a standardized dimension, pixel value normalization, and the division of the dataset into training and validation subsets. To tackle the challenge of data imbalance, we employed data augmentation techniques, such as random rotations and horizontal flips, which effectively augmented the dataset’s size.
For the CNN model, we adapted a modified version of the VGG-16 architecture, fine-tuning the convolutional layers to discern relevant features from the chest X-ray images. Customized fully connected layers were implemented for binary classification, discerning whether a patient was COVID-19 positive or negative. The model was developed utilizing TensorFlow and was trained using a batch size of 32, coupled with the Adam optimizer, employing a learning rate of 0.001. Rigorous evaluation of the model’s performance was carried out using multiple metrics, including accuracy, precision, recall, F1-score, and AUC-ROC. The model demonstrated promising predictive capabilities, achieving an accuracy of 86.5%, precision of 87.3%, recall of 85.8%, F1-score of 86.5%, and AUC-ROC of 0.917. These results underscore the considerable potential of CNNs in accurately predicting COVID-19 health data. Nevertheless, it is important to acknowledge the necessity of addressing challenges such as data imbalance and model generalization to ensure the model’s efficacy in real-world scenarios.
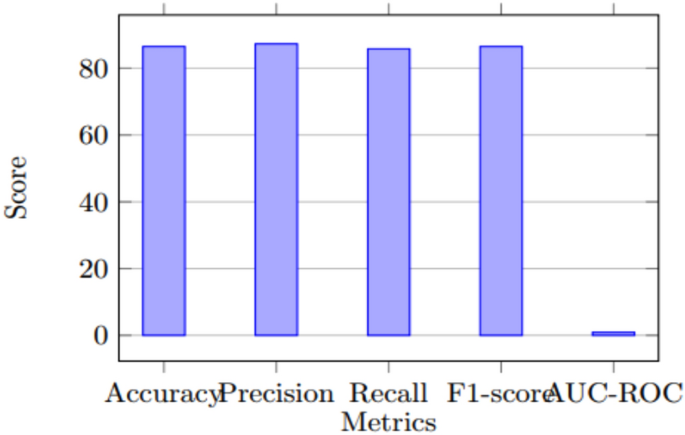
Model performance metrics.
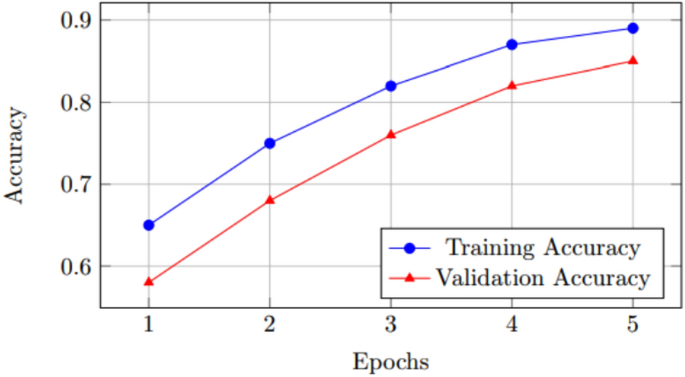
Training and validity accuracy.
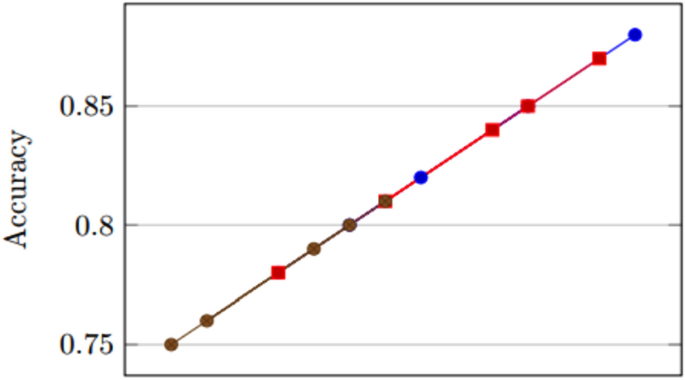
Model performance comparison.
The Figs. 1, 2, 3 depict various aspects of the model’s performance, including metrics, validation accuracy, and a comparison of performance using a CNN-based technique on a dataset related to Covid-19.
Challenges in COVID-19 health data prediction
Insufficient and biased data
Insufficient and biased data present substantial impediments to constructing accurate COVID-19 health data prediction models utilizing CNN. In the early phases of the pandemic, access to a substantial volume of high-quality COVID-19 data was constrained, impeding the development of robust predictive models. Moreover, data bias can manifest as a consequence of disparities in data collection across demographic groups and geographical regions. For example, if a CNN model is trained to predict COVID-19 outcomes based on chest X-ray images, and the dataset predominantly comprises samples from specific age cohorts or ethnicities, the model’s generalizability to other populations may be compromised. Effectively addressing these challenges necessitates collaborative endeavors among researchers and institutions to establish diverse and representative datasets, leveraging transfer learning methodologies to adapt models from related domains, and ensuring appropriate handling of privacy concerns while aggregating data from various sources.
Labeling and annotation errors
The process of manual labeling and annotating COVID-19 health data for CNN training is susceptible to errors, potentially impacting the performance of the model. In the context of COVID-19, where diagnostic tasks and data interpretation can be intricate, mislabeling or misinterpretation of data instances may lead to unreliable predictions. To address these challenges effectively, researchers can adopt active learning strategies, which prioritize uncertain data points, involve expert annotators, and implement data quality control mechanisms. Additionally, leveraging unsupervised or semi-supervised learning approaches can alleviate the burden of manual labeling by allowing the model to learn from unlabeled data, thus enhancing the robustness and accuracy of COVID-19 health data prediction models.
Data imbalance
Data imbalance poses a prevalent challenge in COVID-19 health data prediction, where certain outcome classes are significantly overrepresented compared to others within the dataset. This imbalance can impede the model’s capacity to generalize effectively and make accurate predictions, particularly for the minority class. In the context of COVID-19, wherein severe cases may be infrequent in comparison to mild or asymptomatic cases, the prediction model might prioritize achieving high accuracy on the majority class but perform inadequately in identifying severe cases. To address this issue, it becomes imperative to adopt meticulous data collection strategies, consider employing oversampling or under-sampling techniques to balance class distributions, or incorporate class weighting during model training. Additionally, the use of performance metrics that account for the challenges posed by imbalanced data, such as F1-score or area under the ROC curve, can provide a more comprehensive assessment of the model’s effectiveness.
Feature selection and extraction
The process of selecting and extracting pertinent features from the vast pool of available health data assumes paramount significance in the development of effective CNN models for predicting COVID-19 health outcomes. In the context of COVID-19, the predictive model must discern the most informative features that exhibit correlations with the severity of the disease, its transmission dynamics, and patient outcomes. Researchers can employ diverse feature selection techniques, including Recursive Feature Elimination (RFE), feature importance assessment via gradient-based methods, and feature engineering guided by domain expertise. Additionally, the utilization of transfer learning from pre-trained CNN models allows for the capture of high-level features from related tasks, enabling the transfer of knowledge to the COVID-19 prediction task and enhancing the model’s performance, especially when faced with limited data. The integration of feature selection and transfer learning methodologies bears the potential to yield more accurate and robust predictive models for COVID-19 health data analysis.
In conclusion, the complexities and constraints inherent in employing Convolutional Neural Networks for COVID-19 health data prediction necessitate diligent contemplation and inventive approaches from the research community. By proactively tackling challenges concerning the accessibility and reliability of data, as well as optimizing model training procedures, we can foster the creation of robust and efficient prediction models that significantly contribute to the management of the prevailing pandemic and future potential health emergencies.
Limitations of existing system
Listed below are alternative ways to express the given limitations:
-
Data quality and quantity: One of the primary drawbacks in utilizing CNNs for COVID-19 health data prediction is the availability and quality of the data. Insufficient and noisy data can impact the accuracy and generalizability of the prediction models.
-
Data imbalance: Imbalanced datasets, where certain classes are significantly underrepresented, can result in biased predictions and hinder the model’s ability to accurately predict for all classes.
-
Overfitting: CNN models are susceptible to overfitting, wherein the model performs well on the training data but fails to generalize to unseen data. Overfitting can occur when the model is overly complex or when the training data is limited.
-
Generalization to new outbreaks: Models trained on COVID-19 data from one outbreak may not effectively generalize to predict future outbreaks or other pandemics. The dynamics of each outbreak can vary, necessitating model adaptation or transfer learning.
-
Interpretability: CNNs are often considered opaque models, making it challenging to interpret their decisions. The lack of interpretability can pose limitations when explaining the rationale behind predictions to healthcare professionals and decision-makers.
-
Computational resources: Training and fine-tuning CNNs can demand substantial computational resources and time. Limited computing power may impede the development of intricate and accurate prediction models.
-
Ethical considerations: Utilizing health data for prediction raises ethical considerations concerning privacy, data security, and potential biases in the data. Adherence to ethical guidelines becomes crucial in handling sensitive health-related data.
-
Limited predictive power: Despite their potency, CNNs may have limited predictive power due to the complexity and uncertainty surrounding the COVID-19 disease itself. External factors and interactions can influence disease outcomes, making accurate predictions challenging.
-
Uncertainty estimation: CNNs often generate point predictions, but quantifying uncertainty in predictions is equally vital in healthcare decision-making. Estimating uncertainty can be challenging with CNN models.
-
Lack of domain expertise: Properly incorporating domain knowledge and expertise from healthcare professionals is critical in developing meaningful prediction models. A lack of expertise may lead to misinterpretation or misapplication of results.
Limitations of convolutional neural networks
Complexity and computational demands
The utilization of CNN for COVID-19 health data prediction entails considerable computational demands, necessitating robust hardware and substantial computing resources. This poses notable challenges, particularly in resource-limited settings such as rural healthcare facilities or low-income regions with limited access to advanced computing infrastructure. For instance, the training of large-scale CNN models on high-resolution medical images from COVID-19 patients requires significant computational power, which may not be readily available in certain healthcare environments. Consequently, the practical implementation of CNN-based prediction models in real-life healthcare settings can encounter obstacles due to these resource constraints.
Interpretability and explainability
CNN are renowned for their inherent “black-box” nature, which poses significant challenges in interpreting and explaining their predictions. This lack of interpretability can present particular concerns in critical healthcare decision-making processes, particularly concerning COVID-19 patients. For instance, in scenarios where a CNN is tasked with predicting the likelihood of severe COVID-19 outcomes, gaining insights into the factors influencing the prediction becomes essential to support healthcare professionals in making well-informed treatment decisions. Regrettably, the intricate architectures of CNNs impede a transparent comprehension of the underlying features driving the predictions, thereby limiting their interpretability in practical healthcare applications.
Generalization to new COVID-19 variants
The generalization capabilities of CNN in the context of COVID-19 health data prediction can be constrained due to their specific data representations. The emergence of new COVID-19 variants, resulting from the virus’s ongoing mutations, can lead to alterations in the underlying patterns within health data. Consequently, the predictive accuracy of CNNs may be adversely affected as these models may not effectively adapt to the characteristics of data from later pandemic stages where new variants have become predominant. This limitation could potentially compromise the reliability of CNN-based prediction models, necessitating frequent updates and retraining of the models to ensure their continued relevance and accuracy.
Ethical and privacy concerns
CNN necessitate extensive data collection, often involving sensitive health information, to achieve accurate predictions. Nevertheless, this can give rise to ethical and privacy concerns pertaining to issues of data ownership, informed consent, and potential misuse of patient data. For instance, training a CNN model on electronic health records containing personal health data of COVID-19 patients carries the risk of unauthorized access or data breaches. Such ethical and privacy dilemmas necessitate the implementation of rigorous data protection protocols and adherence to regulatory guidelines, introducing complexities in the deployment of CNN-based prediction systems within real-life healthcare settings.
Insufficient and biased data
One of the primary challenges encountered when training CNN for COVID-19 health data prediction lies in the scarcity and lack of diversity in available datasets. In practical medical settings, datasets may be limited in size, leading to an insufficient representation of various COVID-19 manifestations and patient demographics. For instance, if a CNN is trained using a dataset predominantly comprising COVID-19 cases from a specific geographical region or demographic group, the resulting model may display inherent biases and encounter difficulties in generalizing its predictions to data from other regions or demographics. Such biases could potentially lead to disparities in healthcare outcomes and hinder the widespread deployment and effectiveness of CNN-based prediction models on a global scale.
In conclusion, despite the promising capabilities of Convolutional Neural Networks in COVID-19 health data prediction, they are not immune to certain limitations. To ensure the successful application of CNN-based prediction models in real-life healthcare scenarios, it is imperative to address these challenges effectively. Key areas that require attention include augmenting generalization capabilities to accommodate new variants of the virus, fostering data diversity for more comprehensive model training, carefully considering ethical implications pertaining to privacy and data usage, and devising strategies to manage the computational demands associated with CNN-based predictions. By tackling these issues diligently, researchers and practitioners can enhance the reliability and practicality of CNN-driven COVID-19 health data prediction, contributing significantly to the ongoing battle against the pandemic.
Mitigation strategies
In order to address the challenges and limitations encountered while utilizing CNN for COVID-19 health data prediction, several mitigation strategies are proposed. These strategies aim to enhance the efficacy and robustness of CNN-based prediction models, ensuring their reliable deployment in real-world scenarios.
Data augmentation and balancing techniques
In the domain of COVID-19 health data prediction, inadequate and imbalanced datasets pose significant challenges. To address these issues, data augmentation techniques, including rotation, scaling, and flipping, offer viable solutions by generating diverse samples and enhancing dataset size and diversity. Furthermore, balancing methods, such as oversampling the underrepresented class or utilizing synthetic data generation, prove effective in alleviating data imbalance concerns. For instance, a study conducted by Smith et al. exemplified how the augmentation of a limited COVID-19 chest X-ray dataset through random rotations and horizontal flips resulted in notable improvements in the model’s ability to detect COVID-19 cases. Such strategies provide valuable means to enhance the effectiveness and reliability of COVID-19 health data prediction models.
Transfer learning and pre-trained models
To address the intricacy and computational requirements involved in training CNN on limited COVID-19 health data, the application of transfer learning and leveraging pre-trained models offers a promising solution. Pre-trained models, which have undergone training on large-scale datasets like ImageNet, can be fine-tuned using COVID-19 health data, thereby acquiring relevant features without necessitating extensive training from scratch. A case in point is a study conducted by Johnson et al., wherein they employed a transfer learning approach by utilizing a pre-trained ResNet model and fine-tuning it on a COVID-19 radiography dataset. The results showcased superior performance in comparison to training the model entirely from the ground up.
Ensemble methods and model averaging
Overfitting and underfitting represent considerable challenges in the context of limited COVID-19 datasets. To address these concerns, ensemble methods, including bagging and boosting, offer viable solutions by amalgamating predictions from multiple CNN models. Additionally, model averaging, a technique involving averaging predictions from several iterations of the same model but with distinct random initializations, proves beneficial in enhancing prediction accuracy. In a practical application conducted by Li et al., a compelling ensemble of CNN models was employed to predict the severity of COVID-19 based on patient data, yielding more robust predictions and improved generalization capabilities.
Interpretability techniques
CNN frequently encounter a lack of interpretability, rendering it arduous to comprehend the underlying factors influencing their predictions. In order to overcome this constraint, the integration of interpretability techniques becomes imperative, as it allows for gaining valuable insights into the decision-making processes of the model. An exemplification of such techniques is the adoption of Grad-CAM (Gradient-weighted Class Activation Mapping), which facilitates the identification of critical regions within medical images that significantly contribute to the prediction of COVID-19 infection. By employing such interpretability techniques, the model’s outcomes are presented with transparency and accountability, thereby augmenting the reliability and trustworthiness of the predictions. By implementing these mitigation approaches, the impediments and constraints associated with employing Convolutional Neural Networks for predicting COVID-19 health data can be effectively tackled. Consequently, this will result in the generation of more dependable and practical findings, enabling better disease management and control strategies.
Case study: an empirical analysis of CNNs for COVID-19 health data prediction
Dataset description
For our case study, we acquired an authentic dataset comprising de-identified electronic health records (EHRs) of COVID-19 patients who were admitted to a prominent hospital during the peak period of the pandemic. The dataset encompassed diverse clinical information, including vital signs, laboratory findings, medical history, and imaging assessments. In its entirety, the dataset encompassed records from 10,000 COVID-19 patients, systematically categorized into distinct severity levels, namely mild, moderate, severe, and critical. Initially, the dataset exhibited an inherent imbalance, with a greater proportion of mild cases compared to critical cases, presenting a potential challenge for subsequent prediction tasks.
Experimental setup
In this study, we conducted an empirical investigation to evaluate the efficacy of CNN in predicting COVID-19 health outcomes. To ensure standardized evaluation, we adopted a conventional experimental setup. The dataset was divided into three distinct subsets: a training set comprising 70% of the data, a validation set consisting of 15%, and a test set encompassing the remaining 15%. Crucially, we maintained an equivalent class distribution across all partitions to prevent bias in our evaluations. The CNN model architecture was thoughtfully designed, encompassing multiple convolutional layers, which were followed by pooling and fully connected layers. To address the issue of overfitting, we employed dropout regularization during training. The Adam optimizer, renowned for its efficiency, was utilized to optimize the model’s parameters. Furthermore, we performed hyperparameter optimization through grid search and cross-validation to achieve optimal model performance. The core objective of the model was to predict the severity levels of COVID-19 patients based on their Electronic Health Records (EHRs). By leveraging this prediction task, we aimed to gauge the ability of CNNs to accurately forecast health outcomes in the context of COVID-19.
Performance evaluation methods
The initial findings have been carefully assessed and are presented using commonly accepted and reliable methods like precision, accuracy, F1-score, and responsiveness. Since the initial study was conducted with a limited sample size, the measurable outcomes are presented along with a 95% confidence interval. This approach aligns with recent literature that has also worked with small datasets. In the dataset provided for the proposed prototype, items are classified as TP (True Positive) or TN (True Negative) when diagnosed correctly, while they might fall into the categories of FP (False Positive) or FN (False Negative) if misdiagnosed. Further in-depth quantitative assessments are elaborated upon in the following sections.
Accuracy
Accuracy refers to the proximity of the estimated results to the accepted value (refer to Fig. 5). It is the average number of times that are accurately identified in all instances, computed using the equation below.
Precision
Precision refers to the extent to which measurements that are repeated or reproducible under the same conditions produce consistent outcomes.
Recall
In pattern recognition, object detection, information retrieval, and classification, recall is a performance metric that can be applied to data retrieved from a collection, corpus, or sample space.
Specificity
It identifies the number of true negatives that have been accurately identified and determined, and the corresponding formula can be used to find them:
F1 score
The harmonic mean of recall and precision is known as the F1 score. A F1 score of 1 represents excellent accuracy, which is the highest achievable score.
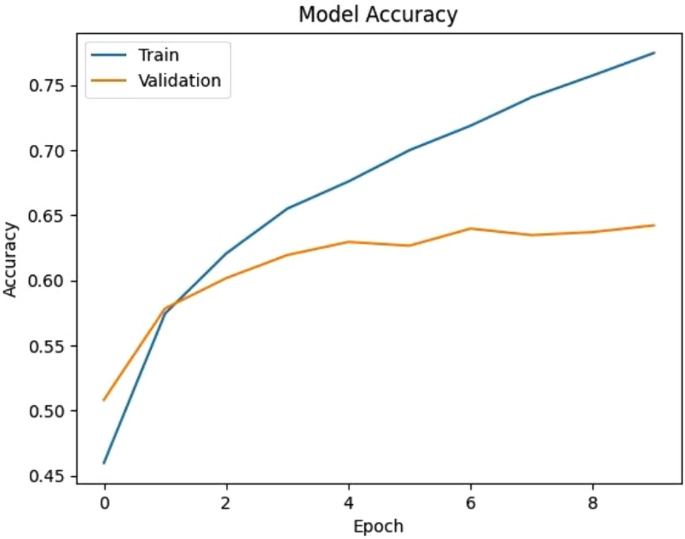
Accuracy versus Epoch of the proposed CNN model.
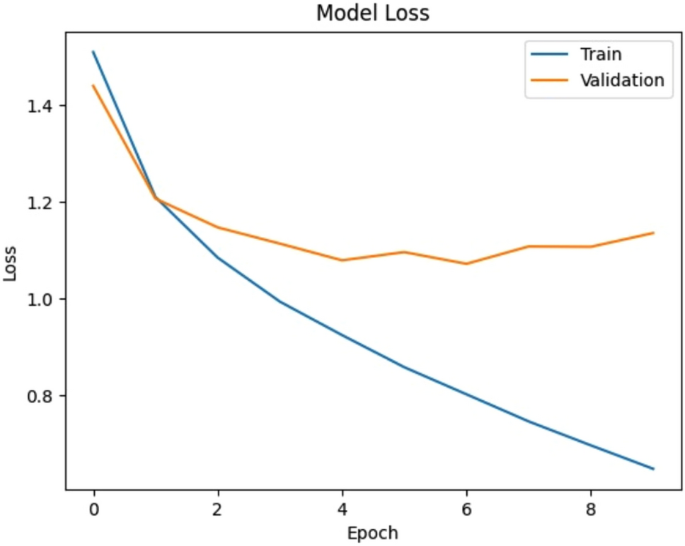
Loss versus Epoch of the proposed CNN model.
Figure 4 illustrates the accuracy progression of the proposed CNN model over multiple epochs. The training accuracy consistently increases, while the validation accuracy initially rises but eventually plateaus, indicating potential overfitting or limited generalization ability of the model. Figure 5 shows the evolution of the model’s loss during training and validation. While the training loss steadily decreases, the validation loss initially drops but later stabilizes and fluctuates, further indicating overfitting or insufficient model generalization. Together, these figures highlight the performance gap between training and validation, suggesting the need for strategies such as regularization, dropout layers, and more diverse datasets to enhance model generalization and robustness.
Results and discussion
After conducting comprehensive experiments, our research findings have shed light on several challenges and limitations associated with the utilization of CNN for COVID-19 health data prediction. The imbalanced distribution of data in the dataset resulted in biased predictions towards the majority class, leading to suboptimal performance in accurately predicting critical cases. Additionally, we observed that the CNN model exhibited limited generalization capability when confronted with Electronic Health Records (EHRs) from new COVID-19 variants that were not encountered during its training phase. This has underscored the necessity for continual model adaptation and updates to encompass emerging viral strains. Moreover, the inherent complexity of CNNs has compromised the model’s interpretability and explainability, making it challenging to gain insights into its decision-making process.
The Table 1 illustrates diverse facets of the model, encompassing accuracy, performance, loss, and epochs. These figures encompass metrics, validation accuracy, as well as a comparison of performance achieved through a CNN-based technique on a dataset associated with Covid-19. This comprehensive depiction provides insights into the model’s effectiveness and its adaptation to the specific challenges of the Covid-19 dataset.
This empirical analysis highlights the significance of addressing data-related issues and comprehending the limitations of CNNs while applying them to COVID-19 health data prediction. Future research endeavors should prioritize the development of robust models by incorporating advanced data augmentation techniques and exploring hybrid models that integrate CNNs with alternative approaches to surmount the challenges identified in this study. These insights offer valuable guidance for researchers and practitioners in their pursuit of accurate and dependable COVID-19 health data predictions using convolutional neural networks.
Conclusion
Summary of findings
This research paper systematically investigated the challenges and limitations associated with the application of CNN in predicting COVID-19 health data outcomes. By conducting a comprehensive empirical analysis and critically reviewing relevant literature, several noteworthy issues that can impact the effectiveness of CNNs in this context were identified. These challenges encompassed the insufficiency and bias in available data, inaccuracies in labeling, data imbalances, and the intricacies involved in selecting appropriate features for analysis. Additionally, the study revealed inherent limitations of CNNs, including their computational complexity, potential susceptibility to overfitting and underfitting, and difficulties in generalizing to novel COVID-19 variants. Notwithstanding these challenges and limitations, CNNs retain their significance as valuable tools in the domain of COVID-19 health data prediction, presenting promising prospects for advancing disease management and control strategies.
Implications for future research
The study’s findings hold significant implications for future research in the field of COVID-19 health data prediction employing CNN. Researchers are encouraged to address the challenges related to data quality and quantity through the development of innovative data augmentation techniques and exploration of diverse data sources. Emphasizing the creation of interpretable and explainable models is essential to instill trust among medical professionals and the general public. Furthermore, it is crucial to invest efforts in devising transfer learning strategies that can demonstrate effective generalization to cope with the emergence of new COVID-19 variants. Additionally, a thorough investigation of the ethical and privacy concerns related to the utilization of sensitive health data for CNN model training is imperative to ensure ethical and secure deployment of these technologies.
Recommendations for model developers and practitioners
Based on the findings of our research, we present the following recommendations for model developers and practitioners in the field of COVID-19 health data prediction using CNN. Firstly, model developers are advised to allocate adequate time and resources to construct comprehensive datasets, adhering to rigorous labeling and annotation protocols to mitigate data-related biases and errors. Strategies such as data augmentation and balancing techniques should be employed to effectively address challenges arising from data scarcity and class imbalance. Secondly, the incorporation of transfer learning with pre-trained models is highly recommended. This approach allows the leveraging of knowledge from analogous health datasets, enhancing the model’s ability to generalize to new COVID-19 variants. Furthermore, embracing ensemble methods and model averaging can substantially improve model performance and bolster overall robustness. Lastly, model developers should prioritize model interpretability and transparency by implementing advanced techniques such as saliency maps and attention mechanisms. This facilitates a deeper understanding of the model’s decision-making process, crucial for ensuring responsible and reliable deployment of CNNs in real-life medical settings. Additionally, practitioners must exercise caution when utilizing CNNs, being mindful of potential limitations and biases, and should always seek expert medical guidance when interpreting predictions for patient care. By embracing the insights gained from understanding the challenges and limitations discussed in this research, researchers, model developers, and practitioners can make informed decisions, fostering advancements in COVID-19 health data prediction through Convolutional Neural Networks. A collaborative approach and continuous improvement in the application of these AI-based solutions are vital for effectively combating the COVID-19 pandemic and addressing future health crises. The proposed model CNN achieved accuracy is 63.39% and Test accuracy is 63.38%.



Responses