Foundation models for materials discovery – current state and future directions

Introduction
The story of AI is a story of data representations (Fig. 1). Early expert systems relied on hand-crafted symbolic representations which eventually evolved into task specific, hand-crafted representations for early machine learning applications1. Since hand-crafting a representation can act as a panacea for a lack of data, and capture a large amount of prior knowledge, this approach persisted for many years. As the availability of data grew, and the amount of compute available to apply to the problem grew with it, thoughts turned to more automated, data-driven ways to learn these representations utilizing the newly popular approach of deep learning2,3,4. This approach, whilst bereft of the injected prior knowledge of their hand-crafted cousins, started to address the related issue of the implicit inclusion of human biases. As this approach gained popularity, driven in part by the advent and enthusiastic uptake of GPUs for model training5,6, we saw a paradigm shift in the way data was considered, with significant effort being placed in the collection and curation of large data sets for training deep learning models7,8. Of course, there is a practical, if not fundamental, limit to the number of clean and large data sets which can be used for such tasks; and fundamental questions about the novelty of scientific discovery which can be brought to bear using models where so much data is already known, which brought into sharp focus the need for more generalizable representations. In 2017, the invention of the transformer architecture9, which was then developed into the generative pretrained transformer (GPT) models by OpenAI10,11,12,13, demonstrated that there was a route to generalized representations through the mechanism of self-supervised training on large corpora of text. Philosophically, this model can be thought of as harking back to the age of specific feature design, albeit through the lens of an oracle trained through exposure to phenomenal volumes of, often noisy and unlabeled, data. Through this decoupling, the task of representation learning, which is the most data hungry, is performed once—with smaller fine-tuning target-specific tasks now requiring little—or sometimes even no—additional training.
Whilst materials discovery can be a somewhat more nuanced task than language generation14,15,16, we have seen that techniques and technology developed in the AI realm for language are often transposed and translated to this important task14,17,18. In this perspective, we will chart the current state of foundation models—the general term for the newly evolved class of machine learning models of which large language models (LLMs) are a part—for materials discovery, and reflect on the challenges which should be addressed to maximize the impact of this important development to the scientific community.
Foundation models
The class of AI model commonly referred to as a “foundation model”—of which LLMs are a specific incarnation—is defined as a “model that is trained on broad data (generally using self-supervision at scale) that can be adapted (e.g., fine-tuned) to a wide range of downstream tasks”19. These models typically exist as a base model, which is often generated through unsupervised pre-training on a large amount of unlabeled data. This base model can then be fine-tuned using (often significantly less) labeled data to perform specific tasks. Optionally, this fine-tuned model can also undergo a process known as alignment. In this process, the outputs generated by the model are aligned to the preferences of the end user. In language tasks, this might take the form of reducing harmful outputs not aligned with human values (Fig. 1), whereas for a chemical task this might take the form of generating molecular structures which have improved synthesisability, or chemical correctness. Typically this is achieved through conditioning the exploration of the latent space to particular parts of a desired property distribution. A visual description of how the encoder and decoder tasks interact with a latent space, as well as how models trained from that latent space produce a property distribution is shown in Fig. 2, and we also point the interested reader at the following excellent articles which are dedicated to the subject20,21,22,23.

This timeline shows the evolution from hand crafted symbolic representations to today’s foundation models through major milestones such as the advent of deep learning.
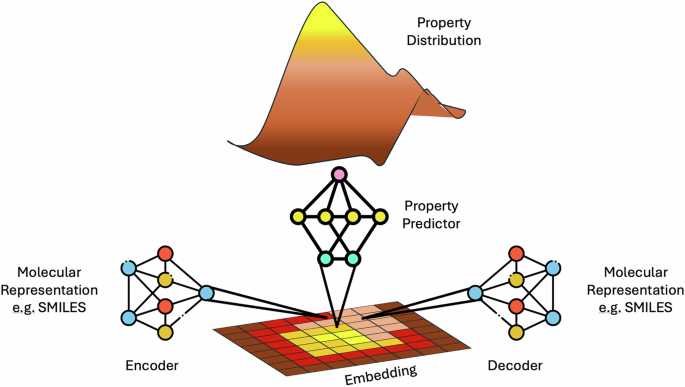
Molecular representations are transformed into their latent space representation by an encoder model, which can be reversed by a decoder model. Additionally properties can be directly predicted from the encoded representation via the building of an additional property predictor model, which is itself capable of producing a property distribution for the latent space.
The separation of representation learning from the downstream tasks such as output generation can naturally be crystallized in the model architecture. Whilst the original transformer architecture encompassed both the encoding and decoding tasks, we now frequently see these components as being decoupled models, leading to encoder-only and decoder-only architectures becoming commonplace. Drawing from the success of Bidirectional Encoder Representations from Transformers (BERT)24, encoder-only models focus solely on understanding and representing input data, generating meaningful representations that can be used for further processing or predictions. Decoder-only models, on the other hand, are designed to generate new outputs by predicting and producing one token at a time based on the given input and previously generated token, making them ideally suited to the task of generating, for example, new chemical entities. Examples of how these types of models could be used in the context of materials discovery are shown in Table 1.
Data extraction
The starting point for successful pretraining and instruction tuning of foundational models is the availability of significant volumes of data, preferably at a high quality. For materials discovery, this principle is even more critical. Materials exhibit intricate dependencies where minute details can significantly influence their properties—a phenomenon known in the cheminformatics community as an “activity cliff”. For instance, in the context of high-temperature superconductors like the high-temperature cuprate superconductors, the critical temperature (Tc) can be profoundly affected by subtle variations in hole-doping levels. Models which do not have the richness of information within their training data may miss these effects entirely, potentially leading to non-productive avenues of research inquiry.
Chemical databases provide a wealth of structured information on materials and are therefore a first choice. Indeed resources such as PubChem21, ZINC25, and ChEMBL26 are commonly used to train chemical foundation models27,28,29. However, these sources are often limited in scope and accessibility due to factors such as licensing restrictions (especially for proprietary databases30), the relatively small size of datasets, and biased data sourcing. Furthermore, one must ensure the quality and reliability of the extracted data. Source documents often contain noisy, incomplete, or inconsistent information, which can propagate errors into downstream models and analyses. For instance, discrepancies in naming conventions, ambiguous property descriptions, or poor-quality images can hinder accurate extraction and association of materials data. To overcome these limitations, there is an imperative need for robust data-extraction models capable of operating at scale on one of the most common and ubiquitous data-sources, that is, the source documents themselves.
A significant volume of relevant materials information is represented in documents, be that public or proprietary scientific reports, patents or presentations. To extract the relevant materials data from these sources, any AI powered extraction models must efficiently parse and collect the materials information from a variety of habitats. Traditional data-extraction approaches primarily focus on text in the documents31,32,33, however, in the realm of materials science, significant information is also embedded in tables, images, and molecular structures. For example, in patent documents, some molecules are selected for their importance and represented by images, while the text can contain any irrelevant structures. Therefore, modern databases aim to extract molecular data not only from text, but from these multiple modalities34,35. Additionally, some of the most valuable data arises from the combination of text and images, such as Markush structures in patents, which encapsulate the key patented molecules. Thus, advanced data-extraction models must be adept at handling multimodal data, integrating textual and visual information to construct comprehensive datasets that accurately reflect the complexities of materials science.
Data extraction-based foundation models typically focus on two types of problems—on the one hand identifying the materials themselves and on the other hand identifying and associating described properties with these materials.
For the former, work to date has focused on leveraging the traditional named entity recognition (NER) approaches36,37 although we note that this is only possible for data encoded within text. Some algorithms have also been developed to identify molecular structures from images in documents, using state-of-the-art computer vision such as Vision Transformers38,39 and Graph Neural Networks40. Recent studies further aim to merge both modalities for extracting general knowledge from chemistry literature41,42,43. For the second type, i.e., property extraction and association, the latest progress in LLMs has allowed such tasks to become much more accurate and leverage schema based extraction44,45.
While traditional NER and multimodal approaches have shown promise in extracting materials data from diverse document formats, it is important to recognize that multimodal language models need not independently handle all forms of information. Instead, they can effectively integrate with specialized algorithms that act as intermediary tools to process specific types of content. For instance, Plot2Spectra46 demonstrates how specialized algorithms can extract data points from spectroscopy plots in scientific literature, enabling large-scale analysis of material properties that would otherwise be inaccessible to text-based models. Similarly, DePlot47 illustrates the utility of modular approaches by converting visual representations such as plots and charts into structured tabular data, which can then be used as input for reasoning by large language models. These examples emphasize that multimodal models can function as orchestrators, leveraging external tools for domain-specific tasks, thereby enhancing the overall efficiency and accuracy of data extraction pipelines in materials science.
Property prediction
The prediction of property from structure is a core component of the value that data-driven approaches can bring to materials discovery. Traditionally, property prediction has either been utilized as a highly approximate initial screen (for example, traditional QSPR methods), or based upon simulation of the fundamental physics of the systems, which can be prohibitively expensive. foundation models offer the opportunity to create powerful predictive capabilities based upon transferrable core components and begin to enable a truly data-driven approach to inverse design.
It is important to note that the current literature is dominated by models which are trained to predict properties from 2D representations of the molecule such as SMILES48 or SELFIES49, which can lead to key information such as the 3D conformation of a molecule, being omitted. This is in part due to the significant delta in available datasets for these two respective modalities—with current foundation models being trained on datasets such as ZINC25 and ChEMBL26 which both offer datasets ~109 molecules—a size not readily available for 3D data. An exception is represented by inorganic solids, such as crystals, where property prediction models usually leverage 3D structures through graph-based or primitive cell feature representations50,51.
Many of the foundation models that are used for property prediction are encoder-only models based broadly on the BERT24 architecture27,52,53,54,55, although Fig. 3 shows that other base architectures such as GPT56,57,58,59 are becoming more prevalent. The reuse of both core models, and core architectural components is a strength of the foundation model approach, although there are analogies to be drawn to the limited number of handcrafted features that ultimately pushed the community into a more data-driven approach.
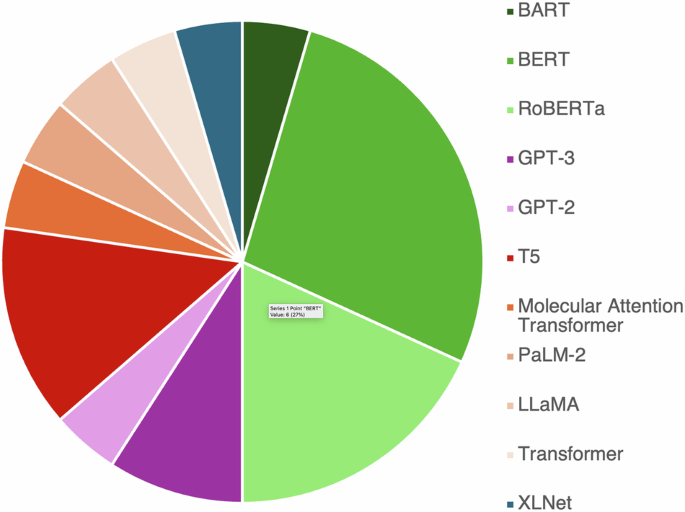
Colors have been determined by the “super type”, or originating, architecture. Greens represent models based upon BERT, purples on GPT, oranges refer to architectures based directly on transformers, and blue XLNet. Data based on ref. 155.
It is also possible to think of some of the recent class of machine learning based potentials—commonly known as MLIPs (machine learned interatomic potentials)—as foundation models60, with their core mode of operation being the prediction of energies and forces of a system, based upon a model pre-trained using a large amount of high-quality reference data—typically based on density functional theory (DFT). In much the same way that base representations can be leveraged to build powerful predictors, we are beginning to see pre-trained models such as MEGNET61, MACE62, ANI63 and AIMNET64,65 being tuned for more specific tasks66, or for more accurate datasets67, for which there is a lower available data volume. This is enhanced by efforts such as Optimade68, which reduces the barrier to bringing together different materials databases, including those built on simulated data.
This offers a fundamentally different route to the use of foundation models for materials property prediction. Whilst many models approach the problem as a direct prediction of a property, this can be hindered by underrepresentation of, for example, rare events. By instead approximating the underlying potential, MLIPs enable traditional simulation techniques at a significantly reduced overhead, and thus the discovery of outcomes which need not be expressed in the original training data.
Of course, for both of these use cases, there is always the problem of understanding transferability and appropriateness of models69,70. To this end, we are beginning to see the emergence of techniques to evaluate properties such as model roughness71,72, which are able to provide quantitative measures which can be related to the likelihood of successful model application and the detection of so-called activity cliffs. We do note, however, that this is still not a solved problem and welcome further research into this area.
Molecular generation
Motivated by the need to overcome the limitations of traditional heuristic and grid-search approaches, AI generative models for material design have gained increasing popularity.
These models are trained to propose novel molecules with desired properties by relying on a variety of molecular structure representations, e.g., text-based SMILES48 and SELFIES49 or graph-based approaches73.
The field of machine learning-based algorithms for designing materials has seen a surge in diversity, with various techniques and applications being proposed. These include VAEs, GANs, GNNs, Transformer-based models, and Diffusion models, each offering unique contributions to the field.
From the end of the last decade, a series of seminal works74,75,76,77,78 based on textual and graphical representations of molecular entities, have started showing promising application of deep generative models to design materials.
In the way paved by these early efforts, prominent examples have demonstrated how these models can be applied to conditionally design and optimize therapeutics successfully validated in silico or in vitro for a variety of applications: kinase inhibitors79,80, antivirals81,82, antimicrobials83, and disease-specific compounds84,85.
The usage of such machine learning-based approaches to molecule generation is not limited to the pharmaceutical domain but has soon shown remarkable results in the broader field of material discovery for property-driven design, e.g., sugar/dye molecules via graph generation86, small molecules, peptides, and polymers generation leveraging language models87,88, and, semiconductors combining deep learning and DFT89.
The development of models has significantly reduced the barriers to accessing generative algorithms for material design. This is largely due to the release of open benchmarks and specialized toolkits for generative molecular design, which encompass a wide range of methods, including evolutionary approaches and generative models, such as GuacaMol90, Moses91, TDC (Therapeutics Data Common)92,93, and GT4SD94.
Despite the ease of access to these technologies, training generative models at the foundation model scale in material science still presents challenges21. Indeed, only recently have we seen attempts to train generative foundation models exhibit promising results in a multi-task setting that leverages extensive pretraining across various chemical tasks95,96.
Foundation models for materials synthesis
The emergence of foundation models in materials science represents a significant opportunity to revolutionize the synthesis of both inorganic and organic materials. The direct application of foundation models in these domains is still in its early stages, with key developments in the synthesis of both inorganic and organic materials thus far being more closely aligned with traditional machine learning approaches rather than foundation models. Nevertheless, compelling indications in the literature suggest that foundation models will play a pivotal role in the future, making it crucial to carefully analyze these trends to anticipate the forms and characteristics that future foundation models may take as they evolve.
Significant advancements in the synthesis of inorganic materials have been achieved through the application of machine learning and data-driven approaches. Recent studies have highlighted the potential of utilizing novel data sources, such as natural language text from scientific literature97, to predict synthesis protocols for inorganic materials98. For example, word embeddings and variational autoencoders have been used to generate synthesis strategies for perovskite materials99. Additionally, natural language processing techniques have been instrumental in designing novel synthesis heuristics derived from text-mined literature data. When combined with active learning, these heuristics optimize the synthesis of novel inorganic materials in powder form100. These efforts have led to the development of extraction and analysis pipelines for synthesis information from scientific publications, laying the groundwork for comprehensive, high-quality datasets for inorganic materials101 and single-atom heterogeneous catalysis102. While natural language processing techniques have demonstrated significant potential, it is critical to observe that literature itself often presents inherent limitations that can affect the applicability of extracted synthesis recipes. For example, the quality and consistency of reporting in the scientific literature, as noted by David et al.103, can pose significant barriers. These challenges include incomplete data, inconsistent terminology, and insufficient experimental details, which can limit the reliability and generalizability of text-derived synthesis strategies. Addressing these limitations will require improved curation of datasets and the development of more robust processing frameworks to ensure accurate and actionable predictions. Supervised machine learning models have been developed to classify and predict suitable synthesis routes and conditions, such as calcination and sintering temperatures, based on target and precursor materials, outperforming traditional heuristics104. Reinforcement learning has also been successfully applied to predict optimal synthesis schedules, including time-sequenced reaction conditions for the synthesis of semiconducting monolayer MoS2 using chemical vapor deposition75. Recent advancements have leveraged high-throughput thermochemical data and classical nucleation theory, identifying favorable reactions based on catalytic nucleation barriers and selectivity metrics, such as in Aykol et al.105. This method has been validated on well-known compounds such as LiCoO₂, BaTiO₃, and YBa₂Cu₃O₇, showcasing its applicability in identifying both established and unconventional synthetic routes. The integration of such frameworks into foundation models could further enhance their ability to predict and optimize complex synthesis processes by providing structured insights into thermodynamic and kinetic factors, opening up exciting possibilities for leveraging foundation models in both the analysis and prediction of time-series data106, as well as their application to chemical synthesis. The progress made in machine learning for inorganic synthesis underscores the immense potential of foundation models, particularly in expanding the capabilities of LLMs. Recent studies107 have demonstrated the effectiveness of LLMs in predicting the synthesizability of inorganic compounds and selecting suitable precursors. In the long term, foundation models are likely to achieve performance levels comparable to specialized ML models, but with significantly reduced development time and costs. This suggests that the future of material synthesis may increasingly be driven by foundation models tailored to multiple tasks within this domain, making them a powerful tool for advancing the field.
The application of foundation models in the synthesis of organic materials follows a similar trajectory. In the realm of organic synthesis, these models hold immense potential for transforming synthetic pathways and optimizing reaction conditions. Early studies have shown that deep learning models can effectively predict reaction outcomes and retrosynthetic pathways, which are critical to the advancement of organic synthesis. A notable example is the Molecular Transformer model108, the first language model in chemical synthesis, which achieved state-of-the-art accuracy in predicting reaction products by treating reaction prediction as a machine translation problem. Recent advancements in prompt-based inference109 have extended the capabilities of these models, enabling chemists to steer retrosynthetic predictions, thereby providing more diverse and creative disconnection strategies while overcoming biases in training data. The introduction of domain-specific LLMs110,111 underscores the importance of fine-tuning with specialized data and integrating advanced chemistry tools112, significantly enhancing the predictive capabilities for synthetic tasks in organic chemistry. The introduction of the first foundation model95 capable of addressing multiple tasks across both chemical17,108,113,114,115 and natural language domains116 marks a breakthrough, demonstrating that sharing information across these domains can enhance model performance, particularly in cross-domain tasks. Furthermore, the implementation LLMs in organic chemistry, exemplified by recent advancements in training a foundational large-scale model with 15 billion parameters for retrosynthesis prediction and generative chemistry117, highlights the potential of foundation models to extend beyond predictive capabilities, and eventually guide experimental efforts in the laboratory. These advancements underscore the potential for further refinement of foundation models, which could lead to enhanced predictive accuracy and, ultimately, more efficient synthesis of complex organic molecules.
The future application of foundation models in the synthesis of both inorganic and organic materials is particularly exciting when considering their ability to learn from diverse data modalities, including spectroscopic data118, crystallography119, and atomistic simulations60. This capability allows for a more holistic understanding of material behavior, facilitating the design of novel compounds with desired properties. The development of multimodal foundation models, which can simultaneously process and learn from these varied data sources, will represent a transformative approach in the synthesis of both organic and inorganic materials. By capturing complex interactions across different modalities, these models will not only improve predictive accuracy but also enable the generation of new hypotheses for experimental validation. This could lead to the discovery of materials with unprecedented properties, advancing fields such as catalysis, drug design, and energy storage. Moreover, their ability to generalize across different types of data positions them as powerful tools for tackling the intricate challenges in materials science, from understanding reaction mechanisms to optimizing synthesis pathways, all within a unique model.
Challenges and future look
Tackling the data challenge with multi-modal models
In the wake of the rising popularity of LLMs’ applications beyond natural language and the introduction of increasingly powerful foundation models, there has been a surge in interest in multimodal approaches. This has led to the development of vision-language models, such as Flamingo120, LLaVA121, and Idefics122,123 which combine different data types to enhance the perception horizon of foundation models. Alongside modeling approaches, multimodal datasets play a crucial role in the success of such models.
Multiple efforts in building web-crawled datasets combining visuals and textual data have become central in machine learning research, culminating with extensive benchmarks promoting advances in multimodal modeling research, e.g., the Cauldron124 or MMMU125. Besides generic datasets for vision-language modeling, there has been a growing interest in compiling data resources to push the boundaries of foundation model perception. Specifically, in the space of egocentric perception, two notable examples are represented by Ego4D126 and Ego-Exo4D127, which provide rich, diverse, and annotated data for research in human activity recognition and exploration.
Despite the growing popularity of multimodal approaches, their application in material discovery remains limited. This limited application is primarily due to the lack of large-scale, high-quality datasets that cover a wide range of materials and their properties. Nevertheless, in recent years, inspired by the successful application of multi-task prompted training and instruction tuning128, models like Text-chem T595 and Regression Transformer (RT)87,88 have been successfully adopted to solve multimodal tasks to accelerate the design of novel materials exhibiting superior performance to specialized single-task models.
These promising attempts at developing multi-purpose models for materials motivate further and deeper experimentation by combining various data sources in model training. Especially considering the wealth of high-quality data that can be generated by simulations, using the latter to fill gaps in real-world data represents a strategy to build more robust and accurate models.
Capturing new types of data for new types of models
The growing use of generative models for synthetic data generation and data augmentation in the domain of materials discovery remains grounded in data from experimental observations, which is indispensable to support the discovery of novel materials14,16. However, fundamental concerns regarding the reproducibility of experimental findings prevail129, and challenges with respect to effective dissemination of high-quality and findable, accessible, interoperable and reusable (FAIR)130 experimental data cut across scientific disciplines. The lack of reproducibility, in particular, can usually be traced back to experimental data and metadata that is flawed or missing131,132. Additional key challenges are the disparity of data formats and schema that must be addressed in order to achieve data interoperability and standardization133, as well as the diversity of tools used to help digitize and organize experimental data134,135.
While the consolidation and organization of already digitized data may be facilitated by the use of LLMs136, the emergence of multi-modal foundation models capable of processing different data modalities at the same time offers new perspectives for data capture and experimental documentation. For example, the shift from convolutional neural networks (CNNs) to transformer-based foundation models for video and action recognition10 has spawned a series of foundational vision-language models that exhibit proficiency in diverse video recognition tasks137,138,139,140,141. Such models, used to describe and document a wide range of real-world actions, have demonstrated significant gains in performance as exemplified by the increase in top-1 accuracy for action recognition on the Kinetics-400 dataset142,143 from 73.9% in 2016 to 93.6% in 2024144.
So far, the training data for large vision-language models has mostly been restricted to common human activities145, but applications have also been studied in specialized domains such as endoscopic surgical procedures to provide information on surgical gestures, generate feedback for surgeons, and help study the relationship between intraoperative factors and postoperative outcomes146. The study and adaptation of multi-modal foundation models to the transcription of specialized procedures has the potential to alleviate data and metadata capture at the roots of the experimentation process, by automatically converting raw data streams of observations and sensor data to a reproducible transcript in the desired target format using multi-modal generative models. Such data capture could be realized with minimal burden on the experimentalist and implemented to generate documentation for any digital system of record, such as electronic laboratory notebooks (ELNs). This type of novel data capture stands to benefit both experimentalists who generate the data as well as data scientists and theoreticians who rely on the data to train and validate scientific models. While this concept is still nascent and lacking open datasets and benchmarks, the automatic transcription of laboratory procedures in real-time using multi-modal foundation models has already been demonstrated in principle147 and may help standardize the process of documenting manual research, link procedural details with outcomes, facilitate science education and information sharing, and supply more consistent and reproducible data for the next generation of foundation models for materials discovery.
Exploiting the science of approximation through multi-fidelity models
As both our model architectures, and the computational infrastructure on which they run, are able to ingest extreme volumes of data, the risk of data collection biases being transferred into the models increases. Whilst we can observe this to a degree in today’s language models—for example through differential capability in English language models compared to other, less common, languages—we believe that this is a particular risk in materials discovery. We hold this view as the very task of discovery requires stepping into unknown chemistries, which may not be well represented in large volume datasets, which can be dominated by a particular part of chemical space (e.g., organic chemistry) or domain (e.g., drug discovery). We view multi-fidelity models as having a significant role to play in mitigating this risk.
A multi-fidelity model is one which can take as input data collected in several different ways, each with their own acquisition cost and accuracy. An example of this might be collecting molecular quantum-chemical data using a range of quantum mechanical approaches (e.g., semi-empirical methods, DFT, MP2). By being able to use data from a variety of sources, in a similar manner to which multi-modal models can draw data from a variety of modalities, multi-fidelity models are able to mitigate data sparsity concerns.
Multi-fidelity methods are a reasonably recent addition to the machine learning for materials discovery toolkit, including recent advances in their use for optimization148, and property prediction of both molecules149,150,151 and materials152,153,154, and we believe that their extension to the foundation models arena will continue to drive progress.
Conclusion
Foundation models are already showing promise in tackling some of the hardest problems in materials discovery. In this perspective we outline some of the fundamentals of this new area of study, and their applicability to the task of materials discovery. We highlight some areas in which foundation models are already being used to significant impact—namely property prediction, retrosynthesis, and molecular generation—and also look to the future to outline areas which we believe are key to continuing to unlock value. These areas hinge on exploiting the natural multi-modality and multi-fidelity characteristics of materials data through increasingly powerful and elegant modeling approaches. We believe that building from the current state of the art into these areas will unlock a significant value stream, potentially enabling substantial acceleration of materials discovery by leveraging the ever-growing troves of data produced by the research community.
Responses