How do Gram-negative bacteria escape predation by Bdellovibrio bacteriovorus?
Introduction
Bdellovibrio bacteriovorus is a small predatory bacterium belonging to the class Bdellovibrionia. Originally isolated from the soil, it was discovered and described by Stolp and Petzold in 19621. They reported the isolation of a predatory microorganism requiring Gram-negative host cells for propagation, with the predator’s life cycle culminating in lysis of the host. Since the 1960s, advances in microscopy, molecular biology and genomics have contributed to a deeper understanding of the lifestyle of this predatory bacterium and have helped shed light on the complex series of interactions required for B. bacteriovorus to locate, recognise, enter, replicate within, and exit its prey.
We now understand that B. bacteriovorus has a complex and tightly regulated bi-phasic lifestyle, which includes a free-swimming or gliding ‘attack phase’ in which predators actively search for prey cells and a ‘growth phase’ which is initiated following successful encounter, recognition and entry into the periplasm of a suitable Gram-negative prey cell2. Once established in the prey periplasm, B. bacteriovorus uses peptidoglycan endo- and transpeptidases to create a spherical, osmotically stable structure known as the bdelloplast in which to replicate3,4. Filamentous growth of the predator is sustained by the degradation of prey-derived macromolecules which are metabolised following the release of diffusible hydrolases into the cytoplasm of the prey. On exhaustion of prey metabolites, synchronous septation of the growing filament occurs which can result in an even or odd number of progeny. Targeted destruction of the prey cell wall during predator exit is facilitated by the deacetylation of prey peptidoglycan and the release of a lysozyme which is specific for this altered substrate5,6. A thorough contemporary overview of the key genes involved in the predatory lifecycle has been published by Caulton and Lovering7. Although frequently described as an obligate intracellular predator, B. bacteriovorus can also exist in a rare ‘host-independent’ form due to mutations primarily associated with the host interaction locus8,9,10.
B. bacteriovorus has the capacity to invade and kill a wide range of Gram-negative bacteria11, including antimicrobial-resistant (AMR) isolates12,13 and potential biothreat agents14,15. Globally, 4·95 million deaths were estimated to be associated with bacterial AMR in 2019, with 1.27 million deaths directly attributable to AMR16. Of particular concern are antibiotic-resistant strains of Gram-negative bacteria belonging to a group known as the ESKAPE pathogens (comprising of Enterobacter genus, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Escherichia coli). These pathogens represent the greatest risk to human health and escape antimicrobial treatment through a number of resistance mechanisms17. Within the EEA/EU areas, E. coli (53.1% of all isolates), K. pneumoniae (34.3%) and P. aeruginosa (18.7%) have high levels of resistance to at least one group of antimicrobials, with multidrug-resistant strains representing 5.1%, 21.2% and 12.6% of isolates, respectively18. The pressing issue of AMR and the need for novel strategies to address this crisis has resulted in several investigations exploring the therapeutic potential of B. bacteriovorus in a range of settings. Research has shown that B. bacteriovorus can reduce the bacterial burden in biofilms13,19,20,21, human serum22 and in a range of in vivo models23,24,25,26. Importantly, a number of reports have now highlighted the potential for predatory bacteria to rescue animals from otherwise lethal bacterial infections. These studies have included the use of B. bacteriovorus to treat lethal Shigella flexneri infections in zebrafish larvae27 and that treatment of SKH-1 mice with B. bacteriovorus can provide significant protection from a lethal challenge of Yersinia pestis15.
Developing B. bacteriovorus as a credible alternative to antibiotics will rely on optimising dosage forms, understanding delivery routes and defining treatment regimens. Moreover, successful clinical outcomes will require developing an intimate knowledge of all aspects of predator-prey interactions. Great strides have been made in this area, particularly by the labs of Sockett and Lovering, who have identified a number of key components involved in prey recognition28, entry4, remodelling3 and exit6. Equally important will be identifying the conditions under which predation may not be successful. Inefficient predation may occur due to multiple factors including unfavourable environmental conditions such as pH or osmolality29,30 lack of physical access to prey which can occur in specific biofilm architectures at high-density prey cell-packing31 or in mixed species biofilms where spatial organisation of the biofilm can protect specific community members21 and the presence of material or non-prey species which may act as decoys to predation32,33,34. A holistic understanding of predation in medically relevant settings will aid in guiding clinicians on the appropriate use of bacterial predators and when treatment is likely to result in clinical success or failure. To this end, we should strive to identify and characterise hitherto unknown mechanisms and scenarios which result in individual bacteria or prey populations escaping predation. Additionally, some of the currently proposed strategies for escaping predation remain poorly defined with little experimental evidence and would benefit from additional studies.
Recently, several reports have been published that shed light on the intriguing ways that prey can escape predation by B. bacteriovorus. The aim of this review is, therefore, to summarise our current understanding of how prey may escape predation and to identify the potential confines of using B. bacteriovorus in a therapeutic setting. By deepening our understanding of all aspects of bacterial predation, we can move towards translating bacterial predators from ‘bench to bedside’ with greater confidence.
Escaping predation
Prior to discussing how prey may escape predation, it is important to highlight that there are still no known simple genetic mutations that result in resistance to predation. Bacterial predation by B. bacteriovorus is a multifactorial process, relying on the action of a diverse range of enzymes acting on multiple targets. This is consistent with the fact that resistance to predation by genetic alteration of a single target has not been reported.
Plastic phenotypic resistance
Predator-prey relationships are observed at all trophic levels of life. Multiple explanations exist for how predator-prey populations are maintained without the extinction of the prey population. Lotka and Volterra proposed a simple model to explain the equilibrium achieved between predator and prey populations35,36. Their model suggests that when predator numbers are low, prey can increase in number. An increase in the prey population results in an abundance of food for predators and thus an increase in predator numbers. However, a larger predator population will drive down the prey population. As food becomes sparse, the predator population will decrease and the cycle repeats. The cyclic nature of this model results in oscillations of the predator-prey populations without extinction of the prey. In terms of predation by B. bacteriovorus, these oscillations are observed under laboratory conditions and prey are never driven to extinction, even in the presence of a high concentration of predators37,38. Previous work suggested that prey populations survived at low densities due to the low probability of the predator finding a prey cell and that these findings fit well with the Lotka-Volterra model39. However, experiments performed by Shemesh & Jurkevitch40 suggest that cells which survive predation and are present at low densities exhibit a transient resistance which allows them to persist in the presence of high predator numbers. Using Erwinia carotovora ssp. carotovora and Escherichia coli as prey, the authors isolated prey cells which survived initial predation (R cells) and examined their susceptibility to subsequent predation. To prove the survival of these cells was not simply a function of low cell density, as previously suggested, the cells were concentrated and exposed to predatory bacteria at high prey density. The authors found that compared to ‘naïve’ prey cells, which had not previously been exposed to the pressure of predation, the R population remained stable whilst naïve prey were rapidly reduced in number. It was also shown this effect is not confined to a single predator, R cells exposed to different bacterial predators (a second B. bacteriovorus strain and Bacteriovorax stolpii) were also resistant to predation. This resistance was determined to be transient in nature, removal of the predator and outgrowth of the previously resistant prey returned populations to a sensitive phenotype. The authors suggest resistance cannot be due to a simple mutational event, as reversion to a sensitive phenotype at population level could not occur as quickly as observed. To confirm this assertion, it would be beneficial to sequence the genomes of susceptible and resistant populations to rule out any mutational events. Instead, it is hypothesised that plastic resistance may arise from the release of predator-derived lytic enzymes during ineffective prey cell encounters. The authors suggest that low levels of these enzymes in the environment may trigger a modification of the prey cell surface or possible mechanical changes to the prey cell envelope. However, experimental evidence for this theory is currently lacking and further studies of these transiently resistant populations may identify alternative mechanisms for this resistant phenotype. It is worth highlighting that these predation-resistant cells are distinct from antibiotic persister cells, which have been shown to be sensitive to predation by B. bacteriovorus41.
The regrowth of presumably transiently resistant prey is not confined to experiments performed in simple lab buffers. Studies investigating the action of B. bacteriovorus in human serum have also observed regrowth of the prey population following initial predation22. However, others have reported that human serum inhibits the predatory action of B. bacteriovorus as a result of high osmolality and serum albumin coating the predator30. As transiently resistant populations may protect prey from extinction, it presents a challenge to the therapeutic use of B. bacteriovorus, where complete eradication of a pathogen is the desired outcome. However, in vivo, B. bacteriovorus may synergise with the antibacterial action of the host immune system to clear an infection, as has been previously reported27. Clearly, further studies are required to fully understand the nature of plastic phenotypic resistance. With the cost of next-generation sequencing reducing dramatically, comparative genomic studies should be performed to rule out mutational events in transiently resistant populations. Transcriptional analyses have also proven to be a powerful tool in deciphering predator-prey interactions2,42,43. Such approaches may help shed light on the regulatory pathways and gene expression profiles involved in phenotypic resistance.
Bacterial structures as barriers to predation
The cell envelope of bacteria is a complex multilayered structure. For Gram-negative bacteria, the outermost layer is typically the outer membrane (OM). The OM exhibits remarkable diversity in its composition and structure, varying in terms of lipid, protein, and polysaccharide content. To overcome the structural and biochemical diversity likely to be encountered by bacterial predators during the process of attachment to the OM, B. bacteriovorus appears to have evolved an exquisite solution. Recent work by Caulton et al.28, revealed that Bdellovibrio encodes a family of phage-tail-fibre-like proteins with diverse adhesin domains. These long fibres have been termed mosaic adhesive trimer (MAT) proteins and are expressed on the predator surface. The authors found a total of 21 different MAT proteins encoded by B. bacteriovorus. These fibres were found to be equipped with a diverse range of adhesive tips, potentially enabling bacterial predators to attach to the broad range of prey epitopes they encounter during bacterial predation and invasion. The binding of a MAT protein (Bd2439) to a specific prey cell glycan, derived from Proteus mirabilis, was further shown in vitro. However, single-gene-deletions of Bd2439, and other MAT encoding sequences, were found to have minimal effect on the efficiency of predation in vivo using either P. mirabilis or E. coli prey. These results suggest that there are likely multiple factors and MAT proteins involved in the process of prey recognition. Further in vivo studies, potentially involving predators deleted for multiple MAT genes, may shed further light on their role in determining prey specificity and their potential levels of redundancy.
In addition to prey membrane diversity, B. bacteriovorus may be faced with the challenge of traversing additional structures exterior to the OM. These can include bacterial capsules and surface layers (S-layers). Bacterial capsules are typically polysaccharide or polypeptide structures which protect bacteria from a range of insults including desiccation44, antimicrobials45, host immune responses46 and bacteriophage47. The biochemistry of capsules is complex, capsule structure can be altered by the position of glycosidic / peptide bonds, isoforms of components and further modifications such as O-acetylation48. This diversity results in significant structural variations between bacterial species and even within bacterial species. For example, E. coli produces approximately 80 distinct capsule types49. Despite the array of capsule types produced by Gram-negative bacteria, there is no direct evidence that these structures provide a barrier to B. bacteriovorus predation. Koval & Bayer were able to show that a capsule-positive and capsule-negative revertant of E. coli K29 were equally susceptible to predation by B. bacteriovorus 109 J as measured by reduction in prey culture turbidity50. Visualisation of predation by transmission electron microscopy revealed several instances where predatory cells had only partially penetrated the capsule and were effectively immobilised at the time of staining. As a number of these events were captured, the authors suggest that penetration of the capsule occurs at a slow enough rate for it to be visualised in progress. The potential slowing effect of predator penetration of the capsule was minimal enough that it did not significantly affect the rate of predation when measured hourly by turbidity compared to the non-capsulated strain. Additional experiments involving the use of time-lapse microscopy to measure prey entry times may reveal if capsules slow penetration of prey cells relative to non-capsulated cells of the same strain. Further studies focused on determining the protection afforded by prey capsules with differing structural chemistries would advance our understanding of this potential obstacle to predation. However, anecdotal evidence suggests that capsules from other species do not outright prevent predation. There are numerous reports in the literature of B. bacteriovorus effectively preying on pathogenic species known to produce a diverse range of capsules including K. pneumoniae22, Serratia marcescens51 and A. baumannii11.
In addition to capsules, some bacteria can assemble a proteinaceous paracrystalline array on their surface, known as an S-layer52. Early work by Koval & Hynes suggested that the freshwater-dwelling Aquaspirillum serpens and Aquaspirillum sinuosum were protected from predation by B. bacteriovorus 109 J by production of S-layers53. The authors proposed that observed predation of specific A. serpens strains was attributable to these isolates producing “patchy” S-layers which did not completely protect the OM, allowing for predator attachment53. However, no experimental data was provided quantifying the integrity of the S-layers produced by these strains. Therefore, further research is required to determine the extent to which S-layers may, or may not, offer protection against intracellular predators. Studies utilising epibiotic predators have revealed that S-layers offer limited protection against these predators. Garcia et al,. first reported that the S-layer of S. marcescens is not sufficient to prevent predation by the epibiotic predator Micavibrio aeruginosavorus54. More recently, time-lapse and cryo-electron microscopy provided stunning images of the epibiotic predator Bdellovibrio exovorus vampirizing Caulobacter crescentus through its S-layer55.
Biofilms and predation
Biofilms are typically defined as a community of bacterial cells attached to a surface and enclosed in an extracellular matrix. This matrix can contain extracellular polymeric substances (such as polysaccharides), DNA, proteins and lipids amongst other components. Similar to bacterial capsules, biofilms are known to protect bacterial communities from harsh environmental conditions and may also prevent antibiotics from reaching their cellular targets56. These structures, therefore, present a potential physical barrier between B. bacteriovorus and its prey. The Kadouri laboratory was the first to establish that biofilms are susceptible to Bdellovibrio predation, reporting that predatory cells could reduce both the biomass and number of viable cells in an established E. coli biofilm19. However, biofilms were observed to offer some protection from predation as planktonic prey cells of the same species were preyed on more efficiently than those in a biofilm19. B. bacteriovorus has also been shown to disrupt monolayer and multilayer biofilms formed by a range of important clinical pathogens11. The factors involved in how biofilms impair predation by B. bacteriovorus is an area of active research. The pioneering work of the Nadell lab has shown that prey cell density, spatial organisation and the composition of bacterial communities within a biofilm can all affect predation by B. bacteriovorus21,31. Biofilm architecture can be highly diverse in terms of both the content of the extracellular matrix and the spatial organisation of bacteria within the biofilm57. Additionally, biofilms can be highly dynamic structures, with the microbial and macromolecular composition of the matrix changing over time58. Using V. cholerae as a model prey species, Wucher et al., investigated how the microheterogeneity of biofilms can impact predation and, in turn, how predation alters the landscape and community assembly of biofilms31. Using high-resolution microscopy, the authors were able to determine that V. cholerae cells on the periphery of a biofilm were susceptible to predation by B. bacteriovorus, whereas cells in the interior of large biofilm clusters appeared to be protected. To determine how the biofilm structure prevents predator cells from entering the interior, the authors dissected images of the biofilm into cubic grids and measured localised amounts of secreted matrix and the density of prey cell-cell packing. This spatial analysis revealed that protection from predation occurs in regions of the biofilm with high matrix accumulation and dense cell-cell packing of V. cholerae prey. The results show that there is a threshold of cell-cell packing, beyond which predators can no longer access their prey.
In addition to growing in monoculture, V. cholerae is known to form dual-species biofilms with E. coli59,60. Recent work, also from the Nadell lab, has focused on how the architecture of these mixed-species biofilms influences predation by B. bacteriovorus21. As discussed in the previous study, V. cholerae can survive within biofilms due to areas of high cell packing. However, E. coli biofilms have been reported to be susceptible to predation11. These findings suggest that predation of a dual E. coli and V. cholerae biofilm might result in clearance of E. coli with mostly recalcitrant V. cholerae clusters. However, using fluorescently tagged predators and prey, Wucher et al., found that E. coli survival under B. bacteriovorus predation increases, whereas V. cholerae survival decreases. This happens when E. coli cells are enveloped within densely packed regions of V. cholerae and thus are protected by the V. cholerae biofilm structure. Concomitantly, a small proportion of the V. cholerae population becomes homogenously mixed with E. coli and no longer develops into the highly packed structure which affords protection from predation, explaining the population level decrease in V. cholerae survival. However, the authors also found that if sufficient E. coli cells are in the immediate vicinity of V. cholerae at the initiation of biofilm formation, then V. cholerae is prevented from establishing a densely packed biofilm structure which can protect both species from predation. The resulting disordered biofilm structure no longer blocks predator entry, resulting in predation of both species.
Some studies also suggest that on encountering biofilms, attack phase cells may upregulate a specific set of genes, including those involved in motility, chemotaxis and proteolysis42,61,62. A review of the anti-biofilm activities of B. bacteriovorus has been produced by Mookherjee and Jurkevitch63.
Biochemical defences to predation
Many Gram-negative bacteria are known to produce a plethora of antimicrobial compounds which may either affect closely related species or be more broadly active. These compounds can include enzymes64, antibiotics65, bacteriocins66 and pyocyanins67. Unsurprisingly, recent studies have reported several prey-derived agents which either directly affect the viability of predators or perturb predation by other mechanisms, such as reducing predator motility. This section will discuss these findings in more detail.
Many bacterial pathogens utilise quorum sensing (QS) to coordinate transcriptional responses based on cell density68. QS plays an important role in bacterial pathogenicity69. It relies on the release of diffusible signalling molecules which accumulate at high cell densities, triggering the expression of specific genes. In pathogenic bacteria, the expression of many virulence factors are controlled in this manner68. As such, QS molecules are likely to be present at the site of infections caused by certain Gram-negative pathogens. Dwidar et al., recently uncovered that one such QS molecule, known as diffusible signalling factor (DSF), is impedes predation by B. bacterivorous 109J70. DSF at environmentally relevant concentrations was shown to slow the predation and lysis of E. coli prey. Additionally, exogenous DSF at a concentration of 50 μM reduced the motility of predatory cells by 50%. Transcriptional analyses revealed that the delayed predation is likely due to the down-regulation of numerous genes important in attack phase (flagellum assembly, chemotaxis, serine proteases) and the up-regulation of stress-related genes.
The results observed for predation in the presence of DSF are similar to those seen previously in the presence of indole71. Indole is also recognised as a bacterial signalling molecule and is commonly produced by members of the gut microbiota72. Physiologically relevant concentrations of indole have been shown to significantly delay predation of both E. coli MG1655 and Salmonella enterica KACC 1159571. As in the previous study, indole was found to significantly reduce the motility of predatory cells and greatly reduce the expression of flagellar assembly genes. As motility is important in facilitating initial predator-prey encounters73, decreased swimming speeds and lack of motility will impact predation kinetics74. Moreover, the addition of indole to prey cells following predator entry was found to slow the release of B. bacteriovorus from bdelloplasts. Microscopic analysis using stained predators and fluorescent prey revealed that the presence of indole delayed both the intraperiplasmic growth of B. bacteriovorus and the lysis of bdelloplasts71.
Others have also reported the anti-Bdellovibrio effects of QS molecules. Hoshiko et al., recently showed that predation-resistant P. aeruginosa PA14 produces quinolone signalling molecules which protect E. coli cells from predation75. However, this anti-predatory effect was found to be diminished when predation assays were performed in the presence of P. aeruginosa mutated for the pqs QS system. The pqs QS system of P. aeruginosa uses the quinolone compounds 4-hydroxy-2-heptylquinoline (HHQ), 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) and 2-heptyl-4-quinolinol-1-oxide (HQNO) as signalling molecules. The addition of exogenous HHQ, PQS or HQNQ to liquid predation assays was found to reduce the predation efficiency of B. bacteriovorious 109 J on E. coli BW25113 prey, with HHQ having the strongest inhibitory effect. The authors show that these compounds are directly bactericidal to B. bacteriovorus, in some cases reducing predator viability by several orders of magnitude compared to controls. Therefore, the anti-predatory action of these QS molecules is likely to be directly attributed to their toxicity towards B. bacteriovorus. The authors suggest the toxic effect of these quinolone molecules may result from their interference with electron transfer reactions. HQNO is known to disrupt electron transfer flow in bacteria, triggering the production of damaging reactive oxygen species76.
QS molecules are not the only prey-derived compounds known to impair predation by B. bacteriovorus. The Mitchell lab reported in 2017 that the production of cyanide by the prey species Chromobacterium piscinae under certain environmental conditions has a detrimental effect on predation77. B. bacteriovorus HD100 was found to prey efficiently on C. piscinae in HEPES buffer but that predation was halted in dilute nutrient broth (DNB). When cultured in DNB, C. piscinae excreted cyanide at concentrations which significantly reduced the motility of attack phase predatory cells and halted the growth of intracellular predators. The resulting ‘stuck’ bdelloplasts did not produce new progeny. Removal of cyanide from prey supernatants by either purging with nitrogen gas or by chelation via the addition of vitamin B12a, restored predation to normal levels, confirming that the inhibitory effect was almost exclusively (if not solely) due to prey-derived cyanide. As other Gram-negative bacteria are capable of cyanogenesis, including important pathogens like pseudomonads78, this mechanism of escaping predation may not be exclusive to C. piscinae and will require further investigation.
Many Gram-negative bacteria are known to excrete a range of proteolytic enzymes which act as virulence factors or are detrimental to competing bacterial species79. The important Gram-negative human pathogen S. marcescens produces extracellular metalloproteases belonging to the Serralysin family of toxins80. These proteases can interfere with predation by the epibiotic predator Micavibrio aeruginosavorus54. Garcia et al. found that S. marcescens deleted for any of the metalloprotease genes (∆prtS, ∆slpB, and ∆ slpE) were more susceptible to predation in comparison to wild-type prey cells. The authors found that secreted metalloproteases did not affect the viability of predators but did reduce predator attachment to prey cells. Additionally, pre-incubation of the prey, but not the predator, with purified metalloprotease was able to block predation, suggesting that the protective activity of the enzyme is due to modification of prey cell envelope components required for predator attachment. Although metalloprotease production did not have any effect on predation by B. bacteriovorus, the presence of other proteases, such as proteinase K, are known to block predation of prey by B. bacteriovorus20. Given the extensive array of proteases produced by Gram-negative bacteria, further characterisation of their potential role in protecting against predation by B. bacteriovorus is a valuable pursuit.
Drag force can impede predation
Transposon-insertion sequencing has proved to be a powerful tool for identifying genetic determinants important for bacterial survival under a specific set of conditions. Briefly, the approach relies on creating a saturated mutant library by introducing a randomly inserting transposon into a selected strain with the aim of disrupting every gene in the genome multiple times at different positions. Sequencing of the resulting library can identify genes essential for viability. The mutant pool can also be exposed to a selective pressure to identify genes important for survival under the desired conditions. Duncan et al., applied this approach by creating a saturated transposon insertion library using the prey species Vibrio cholerae81. The mutant library was then exposed to predation by B. bacteriovorus 109 J. By sequencing surviving prey populations they were able to identify candidate genes involved in determining susceptibility or resistance to predation. Perhaps the most significant finding from this genetic screen was that V. cholerae with insertions in the flagellar motor protein gene motY were more susceptible to predation. These mutants still produced a flagellum but were non-motile, indicating that prey motility was important in escaping predation. The authors hypothesised that highly motile prey, like V. cholerae, will exert a drag force on attached predators and this drag force could slow the entry of predators into prey. An alternative hypothesis could be that non-motile prey are more easily attached to. The hypothesis of increased drag force being responsible for reduced predation rates was supported by experimental data showing that if the viscosity of the medium was increased with ficoll or methylcellulose viscosity (and hence the drag force exerted on predators increased), survival rates for motile V. cholerae improved and there was a concomitant reduction in predator invasions. An increase in medium viscosity by the addition of 10% ficoll was found to result in a small but non-significant reduction in predatory attachment to V. cholerae, suggesting that the observed reduction in predation was not solely a result of decreased predator attachment in the high viscosity environment.
Discussion and concluding remarks
Predators and their prey are often described as existing in a co-evolutionary ‘arms race’. In theory, this means that the evolution of features which enhance the ability of the predator to catch its prey should result in a reciprocal evolutionary response in the prey that improves its ability to escape predation. This constant evolution ensures neither population goes extinct and was described by Van Valen in his ‘Red Queen’ hypothesis82. If this hypothesis was true for the interaction of Bdellovibrio with its prey, we would perhaps expect to see a diverse range of specific anti-predation mechanisms or the rapid evolution of prey resistance due to genetic mutations. However, this does not appear to be the case. Most of the studies discussed in this review describe scenarios whereby prey escape predation due to non-specific mechanisms. For example, quorum sensing molecules have a clearly defined ‘primary’ function in the natural lifecycle of bacteria, cyanide production is toxic to a range of other organisms83, existence of prey in biofilms will have benefits additional to reducing predation. For comparison, the evolution of bacteriophage and their hosts much more closely represents the textbook description of the co-evolutionary ‘arms race’. Bacteria have developed a diverse range of antiphage systems, with over a hundred systems now identified84. These systems cover both innate and adaptive immunity, with innate systems classically represented by restriction-modification systems and adaptive immunity characterised by CRISPR-Cas systems which relies on the acquisition of short DNA fragments from invading bacteriophage. These fragments are used to create a ‘memory’ of the infection which allows bacteriophage to subsequently recognise and degrade bacteriophage DNA with sequences matching the acquired fragments85. In addition to these immune systems, the high selective pressure applied by bacteriophage often selects for bacteriophage insensitive mutants. Such mutants often express altered cell surface receptor proteins which are no longer recognised by cognate bacteriophage receptor-binding proteins86,87,88. The apparent lack of predation-specific resistance mechanisms suggests that resistance to B. bacteriovorus is difficult for prey to evolve or acquire.
The aim of this review is not to temper expectations in relation to the therapeutic use of predatory bacteria but to highlight important aspects which will need to be addressed by the research community if using B. bacteriovorus in a medical setting is to be realised. It should therefore be emphasised that prey escaping predation is not the only challenge to developing B. bacteriovorus as a ‘living antibiotic’. Other important aspects also require further investigation. These include environmental factors at the site of infection which may limit predation (pH, serum, osmolality, antibiotics), potential host (patient) immune responses to predatory bacteria, potential impacts to the host microbiota, large-scale predator production / formulation and regulatory approval. Many of these challenges are discussed in detail in other reviews89,90,91.
Although prey can escape predation in a diverse range of scenarios (Fig. 1), evidence for specialised anti-predation strategies remains mostly elusive. However, this does not exclude the potential for the existence of highly specific mechanisms which may have evolved exclusively to protect prey species. There are numerous reports of Gram-negative bacteria which are preyed on inefficiently or seemingly not at all by Bdellovibrio20,92,93,94,95,96. Although we are beginning to understand the factors involved in protecting some of these species, e.g. it has been suggested that Pseudomonas aeruginosa may have multiple layers of defence75, we are still to determine why predation of other bacteria is troublesome to B. bacteriovorus. One possible explanation could be that the majority of studies have been performed using a limited number of B. bacteriovorus strains (typically HD100 or 109 J). Although recent work has shown that B. bacteriovorus adhesins may have the potential to attach to a multitude of prey epitopes, there is likely to be a limit to the number of binding adhesins which can be encoded by any one strain of predator. Identifying other strains of B. bacteriovorus with differing prey ranges or forced evolution of laboratory strains may be an option to overcome this problem. Conducting long-term evolution experiments in the presence of a single target prey strain can result in enhanced predation of a specific prey strain92. However, others have observed no improved killing on long-term incubation with prey which are recalcitrant to predation97. Alternatively, synthetic biology and genetic engineering may allow us to broaden the prey range of B. bacteriovorus now we have identified some of the adhesins possibly involved in prey attachment. Nevertheless, investigating species known to be recalcitrant to predation is clearly a good place to start when searching for potentially novel mechanisms of escaping predation. Understanding these mechanisms and factors which impair predation will hopefully aid the scientific community in developing B. bacteriovorus as a ‘living antibiotic’.
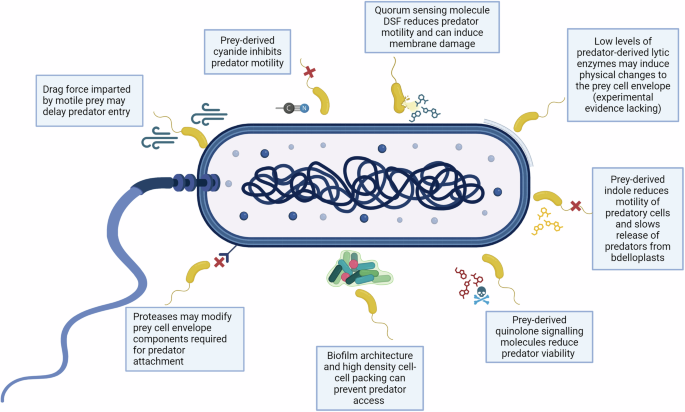
Prey (dark blue) may potentially escape predation by B. bacteriovorus (yellow) via the modification of surface-associated receptors, the production of quorum sensing related molecules (e.g. DSF, quinolone molecules), excretion of chemical compounds that affect predator viability or motility (e.g. cyanide or indole), through the drag force exerted by highly motile prey on attached predators, via plastic phenotypic resistance or due to lack of physical access to prey which can occur in specific biofilm architectures. Figure created with BioRender.com.



Responses