PSMB4: a potential biomarker and therapeutic target for depression, perspective from integration analysis of depression GWAS data and human plasma proteome

Introduction
Depression is a common mental disorders that affects over 300 million people worldwide [1]. with an estimated heritability of 30–40% [2]. Several large scale Genome-wide association studies (GWASs) have been conducted to identify common genetic variants that contribute to depression risk [3,4,5,6]. For example, Wray et al. recruited a major depression GWAS cohort (n = 135,458 cases, n = 344,901 controls) and reported 44 Genome-wide significant (GWS) loci [6]. By extending a broader phenotype definition, Howard et al. reported 102 risk loci for depression in a larger cohort (246,363 cases, 561,190 controls) [3]. but most of these are located in non-coding regions, and the molecular mechanisms underlying their effects are not well understood [3,4,5,6]. Recent advances in multi-omics data integration have provided new opportunities to study the genetic and molecular basis of depression [7,8,9,10]. Dall’Aglio et al. and Li et al. performed transcriptome-wide association study (TWAS) for depression to delineate the gene expression dysregulation respectively [7, 8]. Wingo et al. and Liu et al. conducted proteome-wide association study (PWAS) in human brain tissues to identify protein abundance changes that related depression [9, 10]. These TWASs and PWASs nominated a number of risk genes (e.g. TCTEX1D1, DRD2) and proteins (e.g. CTNND1, RAB27B) for depression which advancing the etiology and therapeutic studies for depression.
However, most of these studies have focused on brain tissues. Human plasma proteome are proteins in the circulation systems which participate in various biological process including immune reaction, signal transduction and etc [11]. Plasma proteome is a major source of human drug target [12, 13], and biomarker for many diseases including psychiatric disorders [14,15,16,–17]. Multiple large-scale human plasma proteome studies had been reported and released in recent years which open new avenues for studying plasma proteome and human complex traits/diseases [15, 18,19,20,21]. Accounting for human plasma proteome is valuable for diseases’ diagnosis, etiology and therapeutic, a survey of human plasma proteome and depression is needed.
In this study, I performed a PWAS analysis by integrating depression GWAS data and human plasma proteome data to identify risk proteins that may be associated with depression risk. I identified four candidate proteins including BTN3A3, PSMB4, TIMP4, and ITIH1. In addition, I found that BTN3A3 and PSMB4 play a causal role in depression, as confirmed by colocalization and Mendelian Randomization (MR) analysis. By compared this plasma PWAS findings and published human brain proteome findings of depression [10, 22, 23], I further confirmed PSMB4 may act as a promising candidate biomarker and therapeutic target for depression.
Materials and methods
GWAS summary statistics
I obtained GWAS summary statistics from Howard et al. [3], which integrated data from three large cohorts (PGC, UK Biobank, and 23andMe) with a total sample size of 807,553 individuals (246,363 cases and 561,190 controls). I downloaded the summary statistics from the Psychiatric Genomics Consortium (PGC) website (https://pgc.unc.edu/for-researchers/download-results/), excluding the 23andMe samples (N = 500,199, including 170,756 cases and 329,443 controls).
Human plasma proteome data
I used a human plasma proteome data from the Atherosclerosis Risk in Communities (ARIC) study [21], which quantified 4657 proteins in 7213 individuals of European American ancestry using modified aptamers (SOMAers). Cis-pQTLs were identified by QTLtools, a total of 2004 significant SOMAers were identified in the European American population.
Plasma proteome-wide association study
I performed a PWAS analysis using FUSION software (http://gusevlab.org/projects/fusion/) [24]. I downloaded plasma PWAS weight files from http://nilanjanchatterjeelab.org/pwas, selecting only the European American population to match the ancestry of the depression GWAS and PWAS weights.
I performed colocalization analysis using the R COLOC package implemented in the FUSION pipeline with the –coloc_P flag. To determine whether the pQTL association signal and GWAS association signal were driven by the same SNP, I utilized COLOC analysis PP4 (post probability of alternative hypothesis 4, with a value between 0 and 1) to indicate whether the two signals are colocalized.
Locuszoom
I used genetic associations between genetic variants and depression extracted from the released depression GWAS summary statistics. Considering the shared genetics between psychiatry diseases [25], I downloaded schizophrenia (67,390 cases and 94,015 controls) and bipolar (41,917 cases, 371,549 controls) GWAS summary statistics respectively to plot the genetic association signals for the target regions [26, 27]. I visualized these associations using the Locuszoom website (http://locuszoom.org/) [28], selecting the European population as the reference population.
Mendelian randomization
I confirmed the causal relationship between the candidate plasma proteins that identified by PWAS using Mendelian Randomization (MR) analysis. I performed two sample MR analysis using TwoSampleMR R package (https://github.com/MRCIEU/TwoSampleMR).
I used human plasma protein QTLs (pQTLs) from the above analysis as instrumental variables in the MR analysis, retaining only pQTLs with association P values under 5 × 10−08 to avoid weak instrument effects. I also performed linkage disequilibrium (LD) clumping when performed MR with default parameters (clump_r2 = 0.001; pop = “EUR”). I applied Inverse variance weighted (IVW) in MR analysis if there were more than one pQTL instruments of a protein while Wald ratio was used if there was only one pQTL instrument for a protein.
Gene and protein expression analysis
I retrieved tissue gene expression data from the GTEx website(https://www.gtexportal.org/) [29], and spatio-temporal gene expression data across human brain development stages from the Human Brain Transcriptome (HBT) website (https://hbatlas.org/) [30]. I queried protein abundance of risk genes on the Human Protein Atlas (www.proteinatlas.org) [31] and performed single-cell gene expression analysis using CoDEx (Cortical Development Expression Viewer, http://solo.bmap.ucla.edu/shiny/webapp/) [32].
Drug-gene interaction analysis
I used DGIdb (The Drug Gene Interaction Database) to explore potential drugs that target the risk proteins I identified in this study [33]. I queried detailed drug information on the DRUGBANK (https://go.drugbank.com/) [34].
Results
Plasma PWAS identified 4 proteins significantly associated with depression
To identify protein abundance alterations that related to depression, I performed a PWAS analysis by integrating depression GWAS and human plasma proteome data. I identified 7 proteins in total at FDR < 0.1, including PSMB4, TIMP4, ITIH1, BTN3A3, ACAA1, SERPING1 and B3GAT3 (Fig. 1; Table 1). Among these proteins, BTN3A3 (P value = 6.41 × 10−06), PSMB4 (P value = 1.42 × 10−05), TIMP4 (P value = 3.77 × 10−05) and ITIH1 (P value = 7.86 × 10−05) are strongly associated with depression (FDR < 0.05). BTN3A3, which is located in the human major histocompatibility complex (MHC) region, showed the most significant association with depression in the PWAS analysis. I also checked the original GWAS association results of the four identified proteins with FDR < 0.05 and found that only genomic loci around the BTN3A3 region reach genome wide significant level while genomic regions around PSMB4, TIMP4 and ITIH1 did not (Fig. 2).
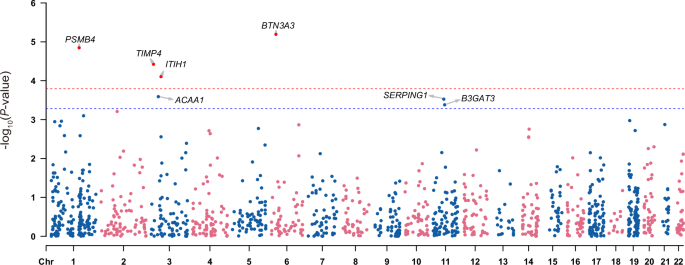
The red dashed line indicates FDR = 0.05 and the blue dashed line indicates FDR = 0.1.
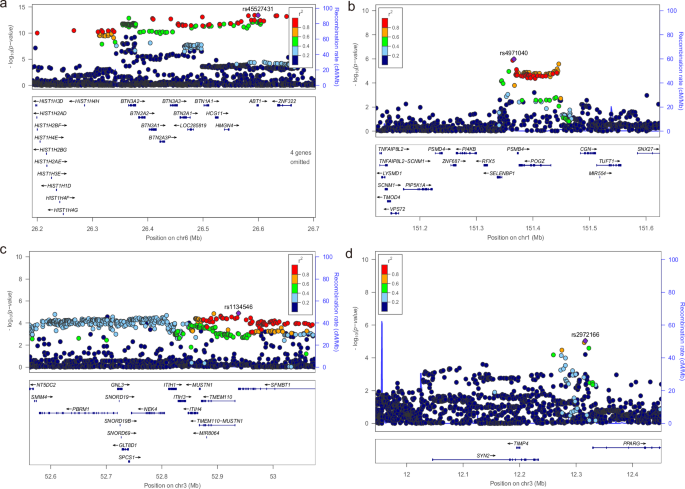
a BTN3A3. b PSMB4. c ITIH1. d TIMP4.
Colocalization analysis indicated BTN3A3 and PSMB4’s causal role for depression
To test whether the genetic association of protein and association of depression are colocalized, I performed colocalization analysis. Among the 7 risk proteins (FDR < 0.1) I identified, BTN3A3 (PP4 = 0.96) and PSMB4 (PP4 = 0.91) showed the highest posterior probability (Table 1). These results indicate that the genetic variants confer risk for depression via affecting protein abundance of BTN3A3 and PSMB4.
Mendelian randomization confirmed BTN3A3 and PSMB4 are causal protein for depression
I performed MR analysis to further confirm the causal relationship between human plasma protein abundance and depression (Fig. 3a). Only the 7 risk proteins I identified in the PWAS analysis were included in further MR analysis. Five proteins, including BTN3A3, PSMB4, TIMP4, ACCA1 and B3GAT3, showed significant association with depression (Table 2). BTN3A3 (MR P value = 9.19 × 10−06) showed the highest association with depression, while PSMB4 (MR P value = 2.65 × 10−04) also showed a significant association with depression (Fig. 3b, c; Table 2). These results, together with the PWAS and colocalization analysis, supported the causal role of protein BTN3A3 and PSMB4 in human plasma with depression.
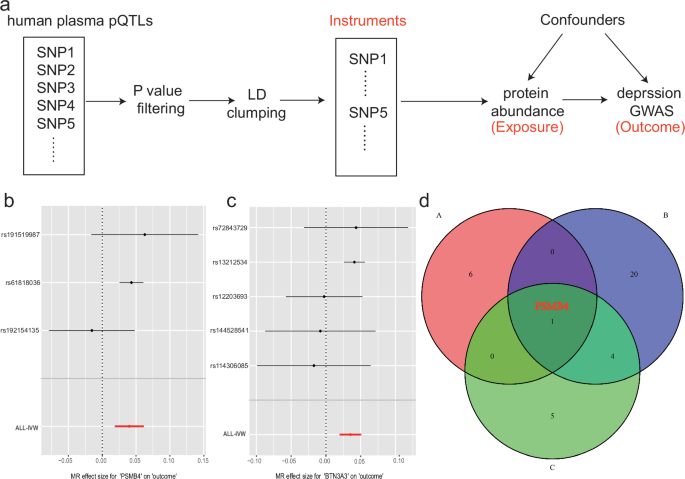
a The overall MR design. b, c The MR analysis result of PSMB4 and BTN3A3. d The comparison between this plasma proteome-wide association study result and published depression proteome-wide association studies and proteome-wide MR studies that using human brain proteome pQTL. A dataset (proteome-wide association study result of this study), B dataset (Wingo et al. [10]), C dataset (Liu et al. [9] and Deng et al. [22]).
Gene and protein expression pattern of BTN3A3 and PSMB4
I further explored the gene and protein expression of BTN3A3 and PSMB4. BTN3A3 and PSMB4 showed a universal expression pattern across the GTEx tissues (Figure S1, S2), and the same pattern was also observed as for protein abundance (Figure S3, S4). BTN3A3 showed an increasing expression during prenatal and stable in postnatal stages while PSMB4 showed a consistent expression level during all brain development stages (Figure S5, S6). PSMB4 is expressed among most of the brain cell types (Figure S9cin Liu et al.) [23] while BTN3A3 is rarely expressed in a specific cell type (Figure S7).
Drug-gene interaction analysis of BTN3A3 and PSMB4
I found 5 drugs (CARFILZOMIB, MARIZOMIB, BORTEZOMIB, IXAZOMIB CITRATE, OPROZOMIB) that had direct interaction with PSMB4, and no result was found for BTN3A3 (Table S1). Among these drugs, CARFILZOMIB, BORTEZOMIB and IXAZOMIB CITRATE are FDA approved drugs used for multiple myeloma treatment, MARIZOMIB was under its Phase 3 clinical trial for Glioblastoma Multiforme (GBM) treatment, and OPROZOMIB was under its Phase 1 clinical trial (Table S1). These results highlight further drug repurposing opportunities for depression treatment targeting PSMB4.
Compare with previous studies further confirmed PSMB4 is a risk protein for depression
Several papers have been published investigating protein abundance in relation to depression. Wingo et al. [10] conducted a PWAS for depression and identified 25 risk proteins, Deng et al. [22] performed a proteome-wide MR analysis for depression and nominated 10 proteins, and Liu et al. conducted an MR analysis focusing on known drug targets for depression and identified 6 risk proteins for depression [23]. These studies primarily focused on integrating depression GWAS and brain proteome data. To determine whether there are robust protein abundance changes between the human plasma proteome and the human brain proteome, I compared the plasma PWAS results with previous studies. These results show that the PSMB4 protein is supported by all these studies, further demonstrating PSMB4 is a promising candidate for depression risk (Fig. 3d).
Discussion
The plasma PWAS analysis aimed to identify aberrant protein abundance changes associated with depression. I identified four plasma proteins (BTN3A3, PSMB4, TIMP4, and ITIH1) that were significantly associated with depression at FDR < 0.05. I further confirmed the causal role of BTN3A3 and PSMB4 in depression using colocalization and MR analysis. A recent plasma proteome pQTL MR study also highlighted the importance of BTN3A3 in three psychiatric disorders, including schizophrenia, bipolar disorder and depression [35]. Additionally, PSMB4 was identified as a transcriptome-wide significant candidate, as reported by Li et al. [8]. Considering the shared genetic between psychiatric diseases [25], I also examined the association of genetic variants near PSMB4 with schizophrenia and bipolar respectively, the lead SNPs for Schizophrenia is rs6684085 (P = 1.88 × 10−05, Figure S8), while the lead SNP for bipolar disorder is rs112766034 (P = 2.22 × 10−04, Figure S9). By comparing this study results with previously published studies for depression that used human brain pQTL datasets, I found convergent evidences supporting PSMB4 as a candidate risk protein in both human brain and blood. This emphasized that PSMB4 may serve as a promising biomarker and potential therapeutic drug target of depression. PSMB4 is a subunit of the 26S proteasome [36], which participate in many biological processes and diseases such as inhibiting cardiomyocyte apoptosis [37], melanoma [38] and breast cancer [39]. I also identified several FDA approved drugs or drugs in clinical trials (e.g. CARFILZOMIB, BORTEZOMIB) that targeting PSMB4, providing further drug repurposing opportunities for depression by targeting PSMB4.
However, this study has several limitations. Firstly, I only focused on the European ancestry proteome and depression GWAS data, and multi-ancestry data should be included in future studies. Secondly, the original study including 23andme cohort, but this data was not available in the current study, and further studies with larger GWAS sample size is needed. Lastly, this study is primarily a computational work, and further experimental validation and exploration are needed to uncover the mechanism of these risk proteins for depression.
In summary, this plasma PWAS analysis identified four plasma proteins significantly associated with depression. Among these proteins, PSMB4 and BTN3A3 may play a causal role in depression based on the coloclization and MR analysis, I also identified drug repurposing opportunities for depression targeting PSMB4. The results provided new potential biomarkers and therapeutic targets for depression and highlight the important role of PSMB4 in depression, which warrants further mechanistic exploration.



Responses